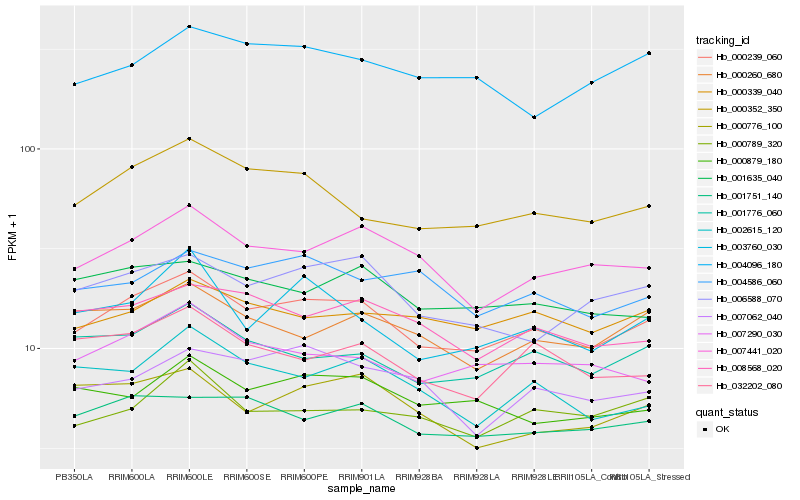
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000239_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007441_020 |
0.0695997778 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 3 |
Hb_000352_350 |
0.0713687446 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001751_140 |
0.0735758493 |
- |
- |
PREDICTED: uncharacterized protein LOC105648510 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001776_060 |
0.0747393875 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 5-like [Jatropha curcas] |
| 6 |
Hb_007290_030 |
0.0747628253 |
transcription factor |
TF Family: bZIP |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 7 |
Hb_006588_070 |
0.0750923215 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 8 |
Hb_008568_020 |
0.0756052516 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 9 |
Hb_001635_040 |
0.0782829906 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 10 |
Hb_000339_040 |
0.0795817619 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 11 |
Hb_000879_180 |
0.0802580276 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 12 |
Hb_000789_320 |
0.0804752497 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000260_680 |
0.080647657 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 14 |
Hb_032202_080 |
0.0809715164 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_003760_030 |
0.0836559448 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 16 |
Hb_007062_040 |
0.0840416762 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 17 |
Hb_004586_060 |
0.0841543777 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 18 |
Hb_002615_120 |
0.0843489613 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004096_180 |
0.0849475897 |
- |
- |
PREDICTED: 14-3-3-like protein A [Jatropha curcas] |
| 20 |
Hb_000776_100 |
0.0854178607 |
- |
- |
Cleavage stimulation factor 50 kDa subunit, putative [Ricinus communis] |