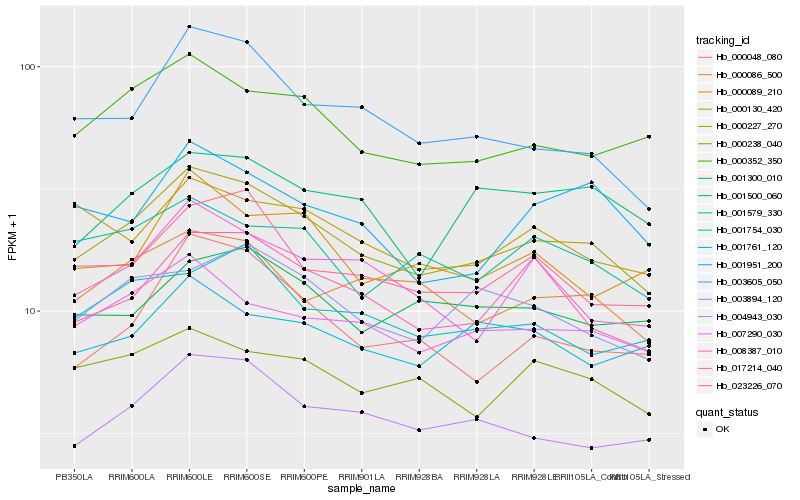
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000227_270 |
0.0 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007290_030 |
0.0716734173 |
transcription factor |
TF Family: bZIP |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 3 |
Hb_000352_350 |
0.0745400183 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |
| 4 |
Hb_017214_040 |
0.0761856589 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase COP1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_023226_070 |
0.0769501804 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004943_030 |
0.0773104243 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |
| 7 |
Hb_000086_500 |
0.081824186 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_000238_040 |
0.0827932682 |
- |
- |
PREDICTED: uncharacterized protein LOC102611758 [Citrus sinensis] |
| 9 |
Hb_003605_050 |
0.0838263804 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_008387_010 |
0.0845313889 |
- |
- |
SMAD/FHA domain-containing protein isoform 3 [Theobroma cacao] |
| 11 |
Hb_003894_120 |
0.0846738888 |
- |
- |
PREDICTED: uncharacterized protein LOC105643110 [Jatropha curcas] |
| 12 |
Hb_001579_330 |
0.085002511 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_001754_030 |
0.0883270946 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 14 |
Hb_000048_080 |
0.0884359548 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000089_210 |
0.0885040582 |
- |
- |
unknown [Medicago truncatula] |
| 16 |
Hb_000130_420 |
0.0891129402 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 17 |
Hb_001300_010 |
0.0907957356 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_001951_200 |
0.0908686943 |
- |
- |
PREDICTED: cold-regulated 413 inner membrane protein 1, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_001500_060 |
0.0913400978 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein ZFN-like [Jatropha curcas] |
| 20 |
Hb_001761_120 |
0.0919785794 |
- |
- |
hypothetical protein JCGZ_22564 [Jatropha curcas] |