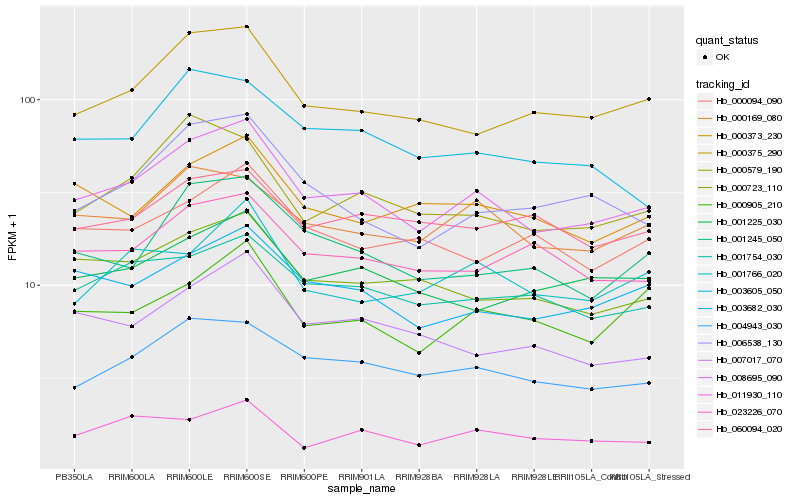
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008695_090 |
0.0 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 2 |
Hb_004943_030 |
0.0770281019 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |
| 3 |
Hb_000375_290 |
0.0795616496 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_003682_030 |
0.079739367 |
- |
- |
hypothetical protein CICLE_v10020876mg [Citrus clementina] |
| 5 |
Hb_000723_110 |
0.0841587023 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 6 |
Hb_001754_030 |
0.0856834805 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 7 |
Hb_000169_080 |
0.0877448142 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 3 [Jatropha curcas] |
| 8 |
Hb_001225_030 |
0.0883747197 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 9 |
Hb_001245_050 |
0.0896744753 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 10 |
Hb_006538_130 |
0.0903251401 |
- |
- |
Uncharacterized protein TCM_009867 [Theobroma cacao] |
| 11 |
Hb_023226_070 |
0.0930680602 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000905_210 |
0.0941227293 |
- |
- |
PREDICTED: protein gar2 [Jatropha curcas] |
| 13 |
Hb_007017_070 |
0.0943375767 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000579_190 |
0.0944525416 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 15 |
Hb_000373_230 |
0.0965962996 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 16 |
Hb_011930_110 |
0.0971426133 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g59720, mitochondrial [Jatropha curcas] |
| 17 |
Hb_003605_050 |
0.0994847791 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_001766_020 |
0.1004978368 |
- |
- |
PREDICTED: probable galacturonosyltransferase 14 [Populus euphratica] |
| 19 |
Hb_060094_020 |
0.1022940426 |
desease resistance |
Gene Name: ATP_bind_1 |
xpa-binding protein, putative [Ricinus communis] |
| 20 |
Hb_000094_090 |
0.1023047626 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |