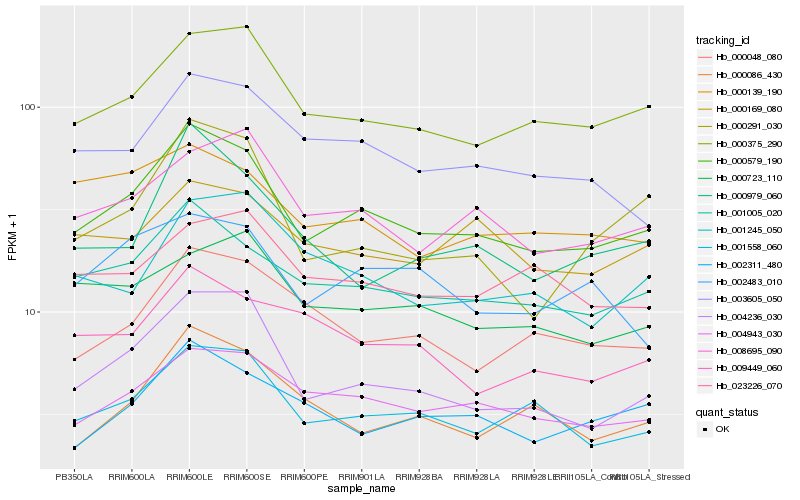
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000579_190 |
0.0 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 2 |
Hb_001005_020 |
0.0729908645 |
- |
- |
PREDICTED: uncharacterized protein LOC105630212 [Jatropha curcas] |
| 3 |
Hb_000375_290 |
0.0786258759 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_001558_060 |
0.0848626888 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004236_030 |
0.0856427324 |
- |
- |
unknown [Lotus japonicus] |
| 6 |
Hb_004943_030 |
0.0860166347 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |
| 7 |
Hb_008695_090 |
0.0944525416 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 8 |
Hb_003605_050 |
0.0985227876 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_002311_480 |
0.1039549413 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000048_080 |
0.1060042227 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002483_010 |
0.1100456182 |
transcription factor |
TF Family: IWS1 |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000979_060 |
0.1101543503 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 2-like [Gossypium raimondii] |
| 13 |
Hb_000291_030 |
0.1104516147 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Eucalyptus grandis] |
| 14 |
Hb_000139_190 |
0.1111377804 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000723_110 |
0.1114832395 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 16 |
Hb_000169_080 |
0.1125105132 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 3 [Jatropha curcas] |
| 17 |
Hb_009449_060 |
0.1132770267 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 18 |
Hb_001245_050 |
0.1142840075 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 19 |
Hb_000086_430 |
0.1143718696 |
- |
- |
hypothetical protein JCGZ_17649 [Jatropha curcas] |
| 20 |
Hb_023226_070 |
0.1151671291 |
- |
- |
conserved hypothetical protein [Ricinus communis] |