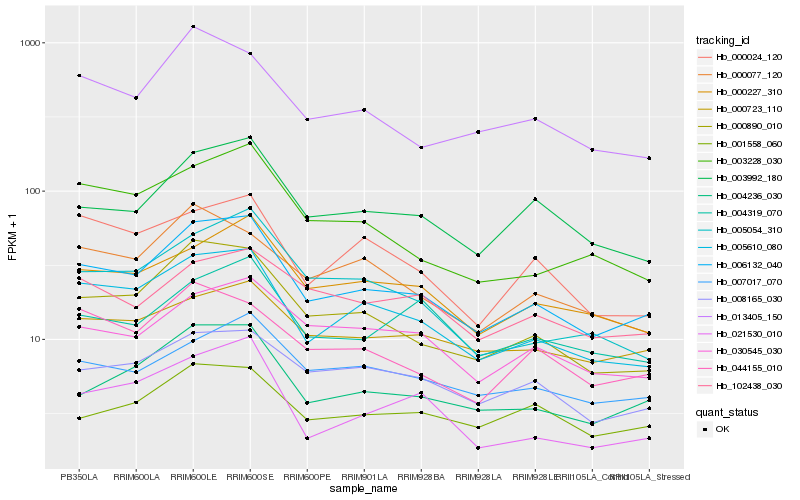
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006132_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_14390 [Jatropha curcas] |
| 2 |
Hb_005610_080 |
0.0697979895 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000890_010 |
0.0765157979 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004236_030 |
0.0805659548 |
- |
- |
unknown [Lotus japonicus] |
| 5 |
Hb_003992_180 |
0.0849167856 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000227_310 |
0.0881188258 |
- |
- |
PREDICTED: cyclin-T1-4-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_008165_030 |
0.0960157828 |
- |
- |
PREDICTED: uncharacterized protein LOC105641288 [Jatropha curcas] |
| 8 |
Hb_030545_030 |
0.0960407867 |
- |
- |
PREDICTED: uncharacterized protein LOC105648743 [Jatropha curcas] |
| 9 |
Hb_007017_070 |
0.0972152808 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_013405_150 |
0.1011237171 |
- |
- |
PREDICTED: actin cytoskeleton-regulatory complex protein pan1 [Jatropha curcas] |
| 11 |
Hb_001558_060 |
0.1033028538 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X2 [Jatropha curcas] |
| 12 |
Hb_044155_010 |
0.103666862 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000723_110 |
0.1047542242 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 14 |
Hb_004319_070 |
0.1049148918 |
- |
- |
hypothetical protein RCOM_0852460 [Ricinus communis] |
| 15 |
Hb_003228_030 |
0.1071269226 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 16 |
Hb_005054_310 |
0.109815708 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 17 |
Hb_102438_030 |
0.1102944692 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000024_120 |
0.1108005386 |
- |
- |
PREDICTED: uncharacterized protein LOC105122546 isoform X1 [Populus euphratica] |
| 19 |
Hb_021530_010 |
0.1108201996 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 20 |
Hb_000077_120 |
0.1110680973 |
- |
- |
PREDICTED: two pore calcium channel protein 1 [Jatropha curcas] |