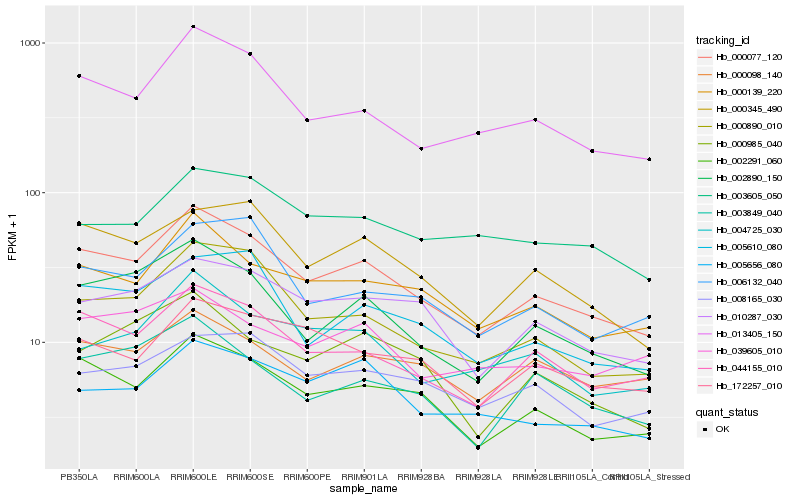
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000077_120 |
0.0 |
- |
- |
PREDICTED: two pore calcium channel protein 1 [Jatropha curcas] |
| 2 |
Hb_002291_060 |
0.070941469 |
- |
- |
PREDICTED: splicing factor 3A subunit 3-like isoform X1 [Fragaria vesca subsp. vesca] |
| 3 |
Hb_044155_010 |
0.0724127875 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002890_150 |
0.0782232444 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 5 |
Hb_013405_150 |
0.0791190652 |
- |
- |
PREDICTED: actin cytoskeleton-regulatory complex protein pan1 [Jatropha curcas] |
| 6 |
Hb_000139_220 |
0.0791638541 |
- |
- |
leucine rich repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_000890_010 |
0.0898581389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000345_490 |
0.0921508904 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 9 |
Hb_010287_030 |
0.0958830534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005610_080 |
0.0983522156 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003849_040 |
0.1048926594 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2-like [Jatropha curcas] |
| 12 |
Hb_172257_010 |
0.1059860052 |
transcription factor |
TF Family: HMG |
high mobility group family protein [Populus trichocarpa] |
| 13 |
Hb_004725_030 |
0.1063532485 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X4 [Jatropha curcas] |
| 14 |
Hb_005656_080 |
0.1076168658 |
- |
- |
PREDICTED: RNA-binding KH domain-containing protein PEPPER-like [Jatropha curcas] |
| 15 |
Hb_008165_030 |
0.1088752927 |
- |
- |
PREDICTED: uncharacterized protein LOC105641288 [Jatropha curcas] |
| 16 |
Hb_000098_140 |
0.1095865008 |
- |
- |
hypothetical protein JCGZ_04989 [Jatropha curcas] |
| 17 |
Hb_006132_040 |
0.1110680973 |
- |
- |
hypothetical protein JCGZ_14390 [Jatropha curcas] |
| 18 |
Hb_000985_040 |
0.1136456204 |
transcription factor |
TF Family: Orphans |
B-box type zinc finger protein with CCT domain isoform 1 [Theobroma cacao] |
| 19 |
Hb_039605_010 |
0.1161578996 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003605_050 |
0.1174021986 |
- |
- |
protein with unknown function [Ricinus communis] |