| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
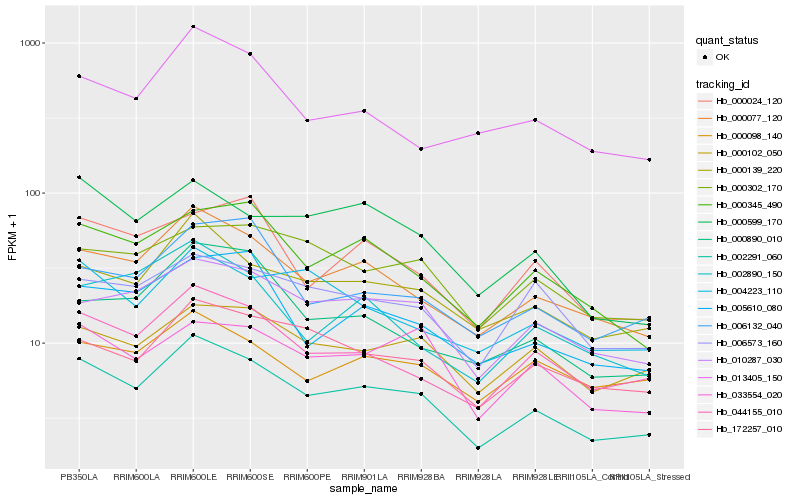
Hb_002291_060 |
0.0 |
- |
- |
PREDICTED: splicing factor 3A subunit 3-like isoform X1 [Fragaria vesca subsp. vesca] |
| 2 |
Hb_000077_120 |
0.070941469 |
- |
- |
PREDICTED: two pore calcium channel protein 1 [Jatropha curcas] |
| 3 |
Hb_044155_010 |
0.082573285 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000139_220 |
0.0852566406 |
- |
- |
leucine rich repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_000345_490 |
0.1002735494 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 6 |
Hb_010287_030 |
0.1007448476 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_172257_010 |
0.1076889401 |
transcription factor |
TF Family: HMG |
high mobility group family protein [Populus trichocarpa] |
| 8 |
Hb_005610_080 |
0.1099034842 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000599_170 |
0.1112134794 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2-like isoform X2 [Populus euphratica] |
| 10 |
Hb_000102_050 |
0.1118565063 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004223_110 |
0.1149035635 |
- |
- |
PREDICTED: putative HVA22-like protein g [Populus euphratica] |
| 12 |
Hb_033554_020 |
0.1157205142 |
transcription factor |
TF Family: BES1 |
PREDICTED: beta-amylase 8 [Jatropha curcas] |
| 13 |
Hb_000024_120 |
0.1161214713 |
- |
- |
PREDICTED: uncharacterized protein LOC105122546 isoform X1 [Populus euphratica] |
| 14 |
Hb_013405_150 |
0.1171208113 |
- |
- |
PREDICTED: actin cytoskeleton-regulatory complex protein pan1 [Jatropha curcas] |
| 15 |
Hb_006573_160 |
0.1185739737 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |
| 16 |
Hb_006132_040 |
0.1187483943 |
- |
- |
hypothetical protein JCGZ_14390 [Jatropha curcas] |
| 17 |
Hb_000302_170 |
0.1193130294 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |
| 18 |
Hb_000890_010 |
0.1201773691 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000098_140 |
0.1205923182 |
- |
- |
hypothetical protein JCGZ_04989 [Jatropha curcas] |
| 20 |
Hb_002890_150 |
0.1207325847 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |