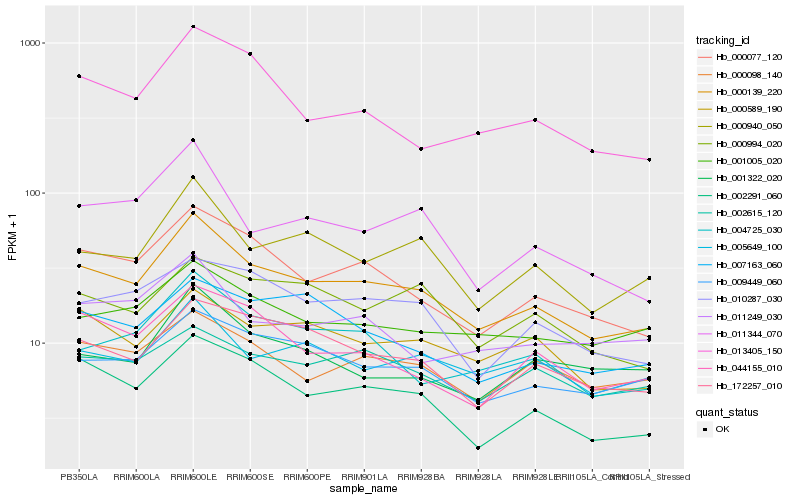
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000139_220 |
0.0 |
- |
- |
leucine rich repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000077_120 |
0.0791638541 |
- |
- |
PREDICTED: two pore calcium channel protein 1 [Jatropha curcas] |
| 3 |
Hb_002291_060 |
0.0852566406 |
- |
- |
PREDICTED: splicing factor 3A subunit 3-like isoform X1 [Fragaria vesca subsp. vesca] |
| 4 |
Hb_000940_050 |
0.0887379257 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 5 |
Hb_009449_060 |
0.0905675301 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 6 |
Hb_172257_010 |
0.094284093 |
transcription factor |
TF Family: HMG |
high mobility group family protein [Populus trichocarpa] |
| 7 |
Hb_000589_190 |
0.0982504454 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004725_030 |
0.0996520373 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X4 [Jatropha curcas] |
| 9 |
Hb_000098_140 |
0.0999095077 |
- |
- |
hypothetical protein JCGZ_04989 [Jatropha curcas] |
| 10 |
Hb_005649_100 |
0.1005283521 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 11 |
Hb_044155_010 |
0.1042030835 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001005_020 |
0.1050458128 |
- |
- |
PREDICTED: uncharacterized protein LOC105630212 [Jatropha curcas] |
| 13 |
Hb_010287_030 |
0.1054578863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_011344_070 |
0.1064699785 |
- |
- |
PREDICTED: non-specific lipid-transfer protein [Jatropha curcas] |
| 15 |
Hb_013405_150 |
0.1074904674 |
- |
- |
PREDICTED: actin cytoskeleton-regulatory complex protein pan1 [Jatropha curcas] |
| 16 |
Hb_007163_060 |
0.1101541767 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 17 |
Hb_002615_120 |
0.1106899437 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 18 |
Hb_011249_030 |
0.1109311291 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 19 |
Hb_001322_020 |
0.1111384158 |
- |
- |
PREDICTED: polycomb group protein VERNALIZATION 2-like isoform X2 [Populus euphratica] |
| 20 |
Hb_000994_020 |
0.1114180368 |
- |
- |
methylenetetrahydrofolate dehydrogenase, putative [Ricinus communis] |