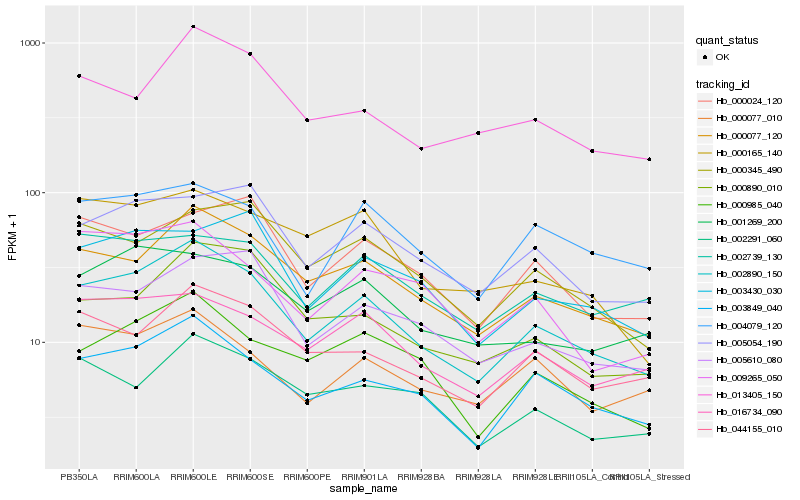
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002890_150 |
0.0 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000077_120 |
0.0782232444 |
- |
- |
PREDICTED: two pore calcium channel protein 1 [Jatropha curcas] |
| 3 |
Hb_003849_040 |
0.0787475434 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2-like [Jatropha curcas] |
| 4 |
Hb_000985_040 |
0.1059797031 |
transcription factor |
TF Family: Orphans |
B-box type zinc finger protein with CCT domain isoform 1 [Theobroma cacao] |
| 5 |
Hb_044155_010 |
0.1063098401 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_005054_190 |
0.110498317 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 7 |
Hb_005610_080 |
0.1115438604 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_016734_090 |
0.1135002092 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein 3-like [Jatropha curcas] |
| 9 |
Hb_013405_150 |
0.1157188249 |
- |
- |
PREDICTED: actin cytoskeleton-regulatory complex protein pan1 [Jatropha curcas] |
| 10 |
Hb_000890_010 |
0.1161025432 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000077_010 |
0.1167535592 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_001269_200 |
0.1197568473 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 13 |
Hb_000345_490 |
0.1206309243 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 14 |
Hb_002291_060 |
0.1207325847 |
- |
- |
PREDICTED: splicing factor 3A subunit 3-like isoform X1 [Fragaria vesca subsp. vesca] |
| 15 |
Hb_004079_120 |
0.1230196899 |
- |
- |
PREDICTED: splicing factor, arginine/serine-rich 19 [Jatropha curcas] |
| 16 |
Hb_003430_030 |
0.1233086517 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 17 |
Hb_000024_120 |
0.1258221318 |
- |
- |
PREDICTED: uncharacterized protein LOC105122546 isoform X1 [Populus euphratica] |
| 18 |
Hb_009265_050 |
0.1258293069 |
- |
- |
PREDICTED: glucosidase 2 subunit beta [Jatropha curcas] |
| 19 |
Hb_002739_130 |
0.1259458904 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Populus euphratica] |
| 20 |
Hb_000165_140 |
0.1295260628 |
- |
- |
PREDICTED: inactive receptor-like serine/threonine-protein kinase At2g40270 isoform X2 [Populus euphratica] |