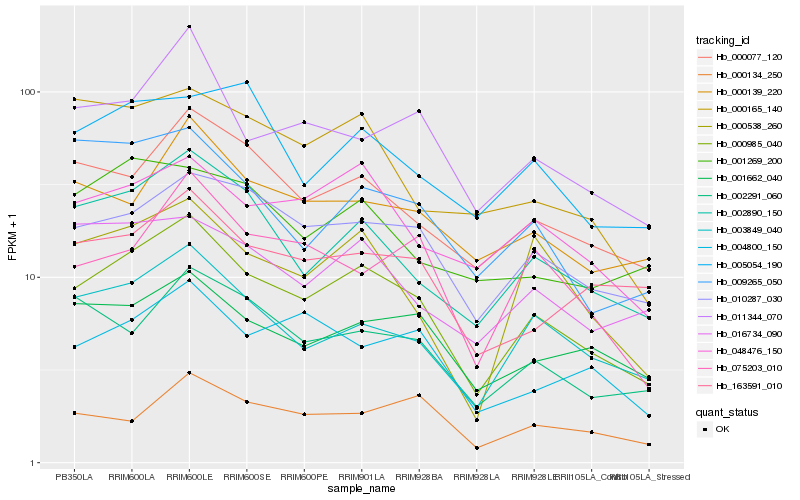
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000985_040 |
0.0 |
transcription factor |
TF Family: Orphans |
B-box type zinc finger protein with CCT domain isoform 1 [Theobroma cacao] |
| 2 |
Hb_003849_040 |
0.1047448468 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2-like [Jatropha curcas] |
| 3 |
Hb_002890_150 |
0.1059797031 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 4 |
Hb_010287_030 |
0.1084473864 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000077_120 |
0.1136456204 |
- |
- |
PREDICTED: two pore calcium channel protein 1 [Jatropha curcas] |
| 6 |
Hb_011344_070 |
0.1234733175 |
- |
- |
PREDICTED: non-specific lipid-transfer protein [Jatropha curcas] |
| 7 |
Hb_048476_150 |
0.1250833953 |
- |
- |
PREDICTED: microtubule-associated protein 70-2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001662_040 |
0.1266157916 |
- |
- |
DNA helicase, putative [Ricinus communis] |
| 9 |
Hb_000538_260 |
0.1300711744 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Citrus sinensis] |
| 10 |
Hb_000139_220 |
0.1320454762 |
- |
- |
leucine rich repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_002291_060 |
0.1328911347 |
- |
- |
PREDICTED: splicing factor 3A subunit 3-like isoform X1 [Fragaria vesca subsp. vesca] |
| 12 |
Hb_004800_150 |
0.1348536133 |
transcription factor |
TF Family: SNF2 |
PREDICTED: ATP-dependent DNA helicase DDM1 [Jatropha curcas] |
| 13 |
Hb_163591_010 |
0.1355188054 |
- |
- |
PREDICTED: amidase 1-like isoform X2 [Populus euphratica] |
| 14 |
Hb_075203_010 |
0.1366685622 |
- |
- |
PREDICTED: calcium-dependent protein kinase SK5 isoform X1 [Jatropha curcas] |
| 15 |
Hb_016734_090 |
0.1385755252 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein 3-like [Jatropha curcas] |
| 16 |
Hb_009265_050 |
0.1425645385 |
- |
- |
PREDICTED: glucosidase 2 subunit beta [Jatropha curcas] |
| 17 |
Hb_000134_250 |
0.1454801366 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_000165_140 |
0.1459490746 |
- |
- |
PREDICTED: inactive receptor-like serine/threonine-protein kinase At2g40270 isoform X2 [Populus euphratica] |
| 19 |
Hb_001269_200 |
0.1467717192 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 20 |
Hb_005054_190 |
0.1474647625 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |