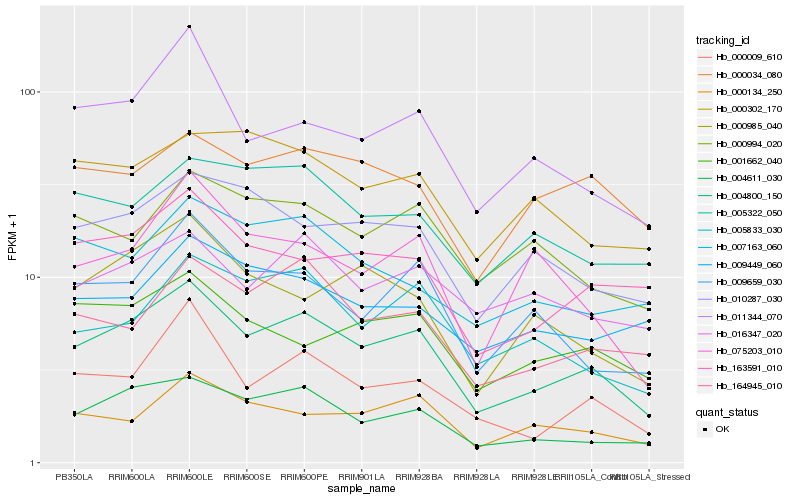
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004800_150 |
0.0 |
transcription factor |
TF Family: SNF2 |
PREDICTED: ATP-dependent DNA helicase DDM1 [Jatropha curcas] |
| 2 |
Hb_004611_030 |
0.0917234673 |
- |
- |
structural maintenance of chromosomes 5 smc5, putative [Ricinus communis] |
| 3 |
Hb_163591_010 |
0.1209445154 |
- |
- |
PREDICTED: amidase 1-like isoform X2 [Populus euphratica] |
| 4 |
Hb_009659_030 |
0.1217627074 |
- |
- |
PREDICTED: beta-galactosidase 17 [Jatropha curcas] |
| 5 |
Hb_164945_010 |
0.1218627459 |
- |
- |
PREDICTED: DNA primase small subunit [Jatropha curcas] |
| 6 |
Hb_000009_610 |
0.1220337116 |
- |
- |
PREDICTED: uncharacterized protein LOC105639638 isoform X2 [Jatropha curcas] |
| 7 |
Hb_011344_070 |
0.1229472706 |
- |
- |
PREDICTED: non-specific lipid-transfer protein [Jatropha curcas] |
| 8 |
Hb_000134_250 |
0.1253744132 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_010287_030 |
0.126695713 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005833_030 |
0.1281627633 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860 [Jatropha curcas] |
| 11 |
Hb_016347_020 |
0.1309435688 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like isoform X1 [Populus euphratica] |
| 12 |
Hb_001662_040 |
0.1315759836 |
- |
- |
DNA helicase, putative [Ricinus communis] |
| 13 |
Hb_000994_020 |
0.1343072108 |
- |
- |
methylenetetrahydrofolate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_005322_050 |
0.1347591739 |
- |
- |
hypothetical protein POPTR_0009s02400g [Populus trichocarpa] |
| 15 |
Hb_000985_040 |
0.1348536133 |
transcription factor |
TF Family: Orphans |
B-box type zinc finger protein with CCT domain isoform 1 [Theobroma cacao] |
| 16 |
Hb_000034_080 |
0.1375243827 |
- |
- |
PREDICTED: MADS-box protein AGL24-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_075203_010 |
0.1378354898 |
- |
- |
PREDICTED: calcium-dependent protein kinase SK5 isoform X1 [Jatropha curcas] |
| 18 |
Hb_009449_060 |
0.1401190154 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 19 |
Hb_007163_060 |
0.1403602716 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 20 |
Hb_000302_170 |
0.1421455311 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |