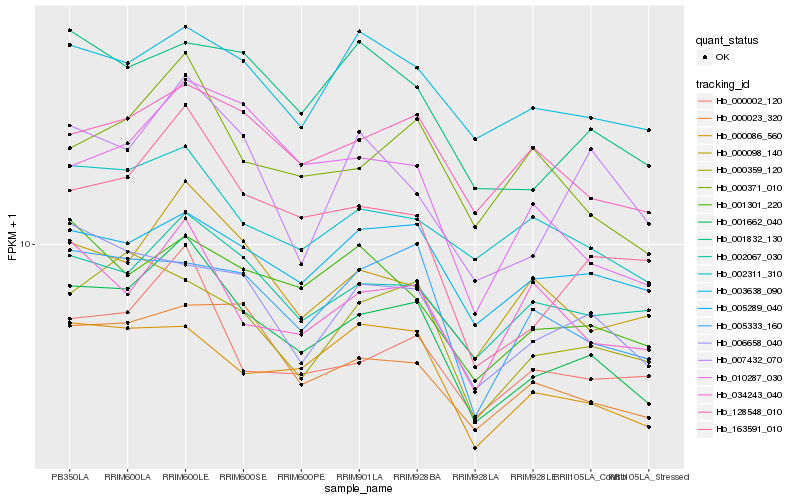
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001662_040 |
0.0 |
- |
- |
DNA helicase, putative [Ricinus communis] |
| 2 |
Hb_163591_010 |
0.0817324404 |
- |
- |
PREDICTED: amidase 1-like isoform X2 [Populus euphratica] |
| 3 |
Hb_000002_120 |
0.0913320402 |
- |
- |
PREDICTED: uncharacterized protein LOC105782064 [Gossypium raimondii] |
| 4 |
Hb_000023_320 |
0.0975707415 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002067_030 |
0.0980121238 |
- |
- |
PREDICTED: poly(ADP-ribose) glycohydrolase 1-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_000371_010 |
0.0991006155 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 7 |
Hb_000098_140 |
0.0991885395 |
- |
- |
hypothetical protein JCGZ_04989 [Jatropha curcas] |
| 8 |
Hb_000086_560 |
0.1006229112 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001832_130 |
0.104152764 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 10 |
Hb_002311_310 |
0.105832475 |
- |
- |
PREDICTED: uncharacterized protein LOC100246622 [Vitis vinifera] |
| 11 |
Hb_128548_010 |
0.1072288596 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000359_120 |
0.1077108979 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001301_220 |
0.1082246076 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 4 [Jatropha curcas] |
| 14 |
Hb_010287_030 |
0.108877061 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005333_160 |
0.1099118618 |
- |
- |
hypothetical protein B456_009G230200 [Gossypium raimondii] |
| 16 |
Hb_007432_070 |
0.111455217 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 17 |
Hb_034243_040 |
0.1115451109 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1-like [Jatropha curcas] |
| 18 |
Hb_006658_040 |
0.1118245968 |
- |
- |
hypothetical protein POPTR_0006s19640g [Populus trichocarpa] |
| 19 |
Hb_005289_040 |
0.1127363408 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 20 |
Hb_003638_090 |
0.1134055138 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |