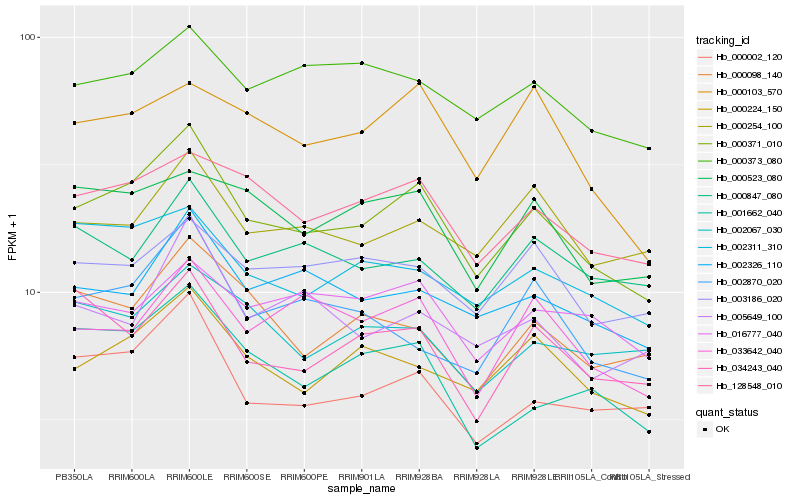
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000371_010 |
0.0 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 2 |
Hb_128548_010 |
0.0820299282 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000224_150 |
0.0822748174 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000002_120 |
0.084761734 |
- |
- |
PREDICTED: uncharacterized protein LOC105782064 [Gossypium raimondii] |
| 5 |
Hb_002326_110 |
0.0912336904 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 6 |
Hb_033642_040 |
0.0953075033 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002311_310 |
0.0954028354 |
- |
- |
PREDICTED: uncharacterized protein LOC100246622 [Vitis vinifera] |
| 8 |
Hb_000523_080 |
0.0982728537 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 9 |
Hb_001662_040 |
0.0991006155 |
- |
- |
DNA helicase, putative [Ricinus communis] |
| 10 |
Hb_000103_570 |
0.1003366752 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000373_080 |
0.1005500726 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 12 |
Hb_000098_140 |
0.1006727274 |
- |
- |
hypothetical protein JCGZ_04989 [Jatropha curcas] |
| 13 |
Hb_000254_100 |
0.1007937519 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 14 |
Hb_003186_020 |
0.1015297891 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 15 |
Hb_034243_040 |
0.1017213104 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1-like [Jatropha curcas] |
| 16 |
Hb_000847_080 |
0.1032345995 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 17 |
Hb_002870_020 |
0.103705801 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005649_100 |
0.1060151486 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 19 |
Hb_002067_030 |
0.1069883228 |
- |
- |
PREDICTED: poly(ADP-ribose) glycohydrolase 1-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_016777_040 |
0.1070992815 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |