| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
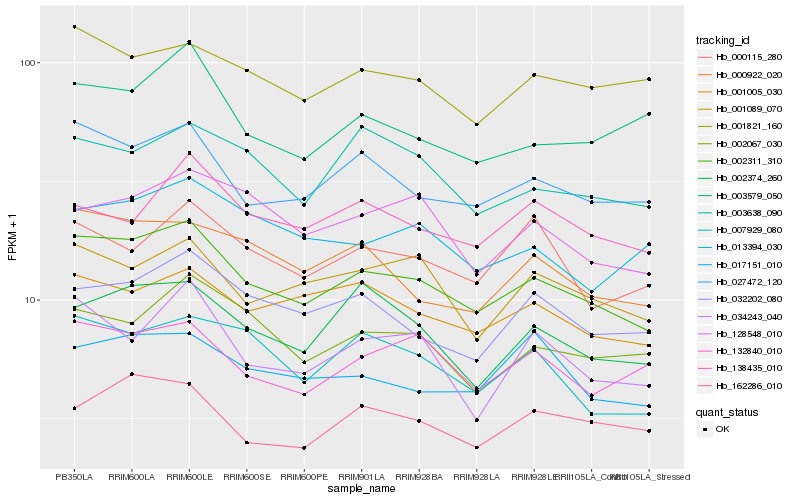
Hb_002311_310 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC100246622 [Vitis vinifera] |
| 2 |
Hb_027472_120 |
0.0597260654 |
- |
- |
PREDICTED: thiosulfate/3-mercaptopyruvate sulfurtransferase 1, mitochondrial isoform X1 [Jatropha curcas] |
| 3 |
Hb_001005_030 |
0.0753078756 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 4 |
Hb_000922_020 |
0.0755570574 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_032202_080 |
0.0774036831 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_001089_070 |
0.0780928955 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 7 |
Hb_007929_080 |
0.0794306352 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 8 |
Hb_002374_260 |
0.0794792749 |
- |
- |
PREDICTED: probable methyltransferase-like protein 15 [Jatropha curcas] |
| 9 |
Hb_002067_030 |
0.0813498145 |
- |
- |
PREDICTED: poly(ADP-ribose) glycohydrolase 1-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_003638_090 |
0.0824012286 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |
| 11 |
Hb_132840_010 |
0.0824931289 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 12 |
Hb_001821_160 |
0.0836288333 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 13 |
Hb_017151_010 |
0.0855441933 |
- |
- |
PREDICTED: uncharacterized protein LOC105641966 [Jatropha curcas] |
| 14 |
Hb_128548_010 |
0.0858409592 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_162286_010 |
0.0859192264 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 16 |
Hb_034243_040 |
0.0859915444 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1-like [Jatropha curcas] |
| 17 |
Hb_013394_030 |
0.0868377939 |
- |
- |
PREDICTED: exocyst complex component SEC6 [Vitis vinifera] |
| 18 |
Hb_000115_280 |
0.0874428872 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 19 |
Hb_138435_010 |
0.0880834704 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 20 |
Hb_003579_050 |
0.0882331589 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |