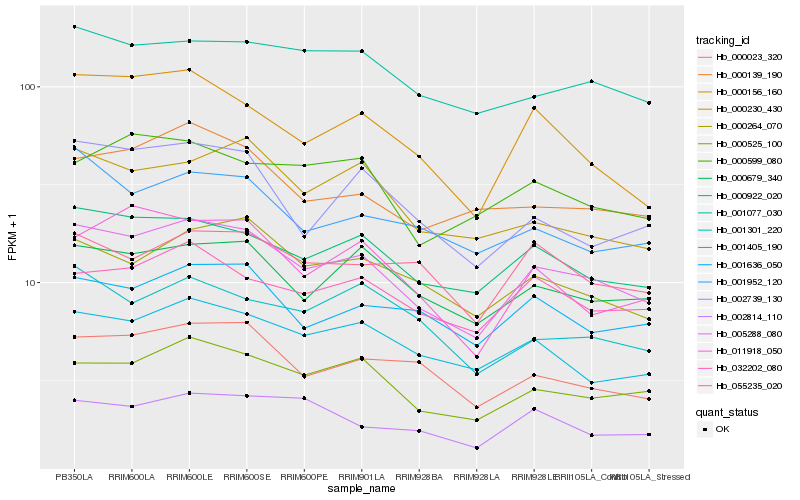
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005288_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000922_020 |
0.0656445704 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_000525_100 |
0.0681784469 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 4 |
Hb_000679_340 |
0.0770648587 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000264_070 |
0.0819649635 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 6 |
Hb_001077_030 |
0.0821749851 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000156_160 |
0.0826800199 |
- |
- |
PREDICTED: urease accessory protein G [Jatropha curcas] |
| 8 |
Hb_001405_190 |
0.0829781015 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000023_320 |
0.0830563022 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_032202_080 |
0.0836132499 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001636_050 |
0.0871326083 |
transcription factor |
TF Family: NF-X1 |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 12 |
Hb_000139_190 |
0.0893305838 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_011918_050 |
0.0909912187 |
- |
- |
- |
| 14 |
Hb_002739_130 |
0.0910412703 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Populus euphratica] |
| 15 |
Hb_000230_430 |
0.0910622758 |
- |
- |
catalytic, putative [Ricinus communis] |
| 16 |
Hb_001952_120 |
0.0924857704 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000599_080 |
0.0933240094 |
- |
- |
PREDICTED: TOM1-like protein 1 [Malus domestica] |
| 18 |
Hb_002814_110 |
0.0940462659 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase TDR [Jatropha curcas] |
| 19 |
Hb_001301_220 |
0.0945790752 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 4 [Jatropha curcas] |
| 20 |
Hb_055235_020 |
0.0955947395 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X1 [Jatropha curcas] |