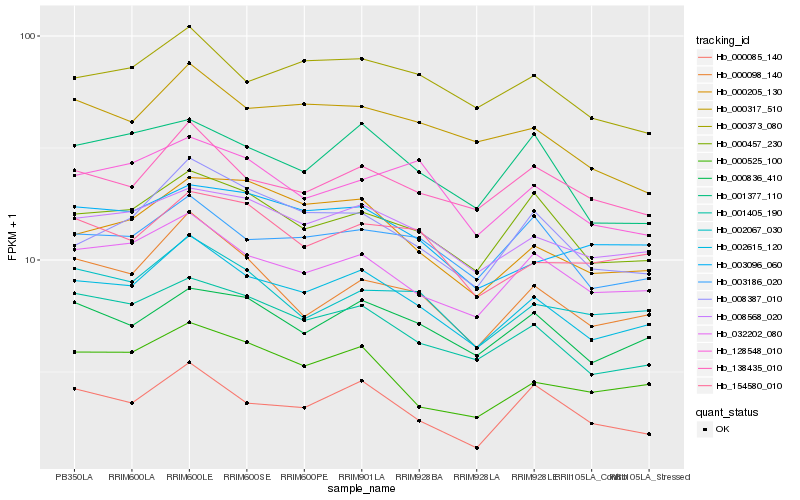
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002615_120 |
0.0 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001405_190 |
0.055365448 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 3 |
Hb_008568_020 |
0.0573790454 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 4 |
Hb_032202_080 |
0.0578358278 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000457_230 |
0.0640190869 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000836_410 |
0.0669249419 |
- |
- |
sec10, putative [Ricinus communis] |
| 7 |
Hb_000098_140 |
0.0677101874 |
- |
- |
hypothetical protein JCGZ_04989 [Jatropha curcas] |
| 8 |
Hb_000525_100 |
0.0693586386 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 9 |
Hb_002067_030 |
0.0704635923 |
- |
- |
PREDICTED: poly(ADP-ribose) glycohydrolase 1-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_008387_010 |
0.0705798648 |
- |
- |
SMAD/FHA domain-containing protein isoform 3 [Theobroma cacao] |
| 11 |
Hb_003186_020 |
0.0706148908 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000317_510 |
0.0721289265 |
- |
- |
PREDICTED: long chain base biosynthesis protein 1 [Jatropha curcas] |
| 13 |
Hb_000373_080 |
0.0724028095 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 14 |
Hb_138435_010 |
0.0727662187 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 15 |
Hb_154580_010 |
0.0729494041 |
- |
- |
hypothetical protein PRUPE_ppa002109mg [Prunus persica] |
| 16 |
Hb_000205_130 |
0.0737795442 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 17 |
Hb_001377_110 |
0.0773310738 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 18 |
Hb_003096_060 |
0.0786294406 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 19 |
Hb_128548_010 |
0.0788989521 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000085_140 |
0.0790003126 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105633025 [Jatropha curcas] |