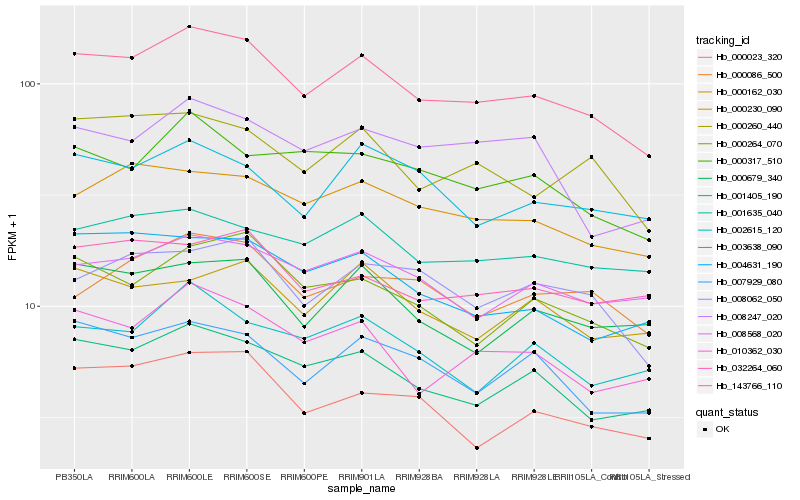
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_143766_110 |
0.0 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 1 [Jatropha curcas] |
| 2 |
Hb_001405_190 |
0.0588834169 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000162_030 |
0.0658389796 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_000260_440 |
0.0699742621 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 [Jatropha curcas] |
| 5 |
Hb_000264_070 |
0.0702557021 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 6 |
Hb_010362_030 |
0.0702696914 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 7 |
Hb_007929_080 |
0.0704396119 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 8 |
Hb_001635_040 |
0.0734949068 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 9 |
Hb_000679_340 |
0.0746467474 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 10 |
Hb_008568_020 |
0.0757242757 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 11 |
Hb_000317_510 |
0.0758088969 |
- |
- |
PREDICTED: long chain base biosynthesis protein 1 [Jatropha curcas] |
| 12 |
Hb_008062_050 |
0.0759827221 |
- |
- |
PREDICTED: uncharacterized protein LOC105631631 [Jatropha curcas] |
| 13 |
Hb_004631_190 |
0.0775960503 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 14 |
Hb_000023_320 |
0.0787669556 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000230_090 |
0.0789018691 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 16 |
Hb_032264_060 |
0.0790189772 |
- |
- |
hypothetical protein JCGZ_02506 [Jatropha curcas] |
| 17 |
Hb_000086_500 |
0.0793442935 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_003638_090 |
0.0804669851 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |
| 19 |
Hb_002615_120 |
0.0818204705 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 20 |
Hb_008247_020 |
0.0830299748 |
- |
- |
PREDICTED: cyclin-L1-1-like [Jatropha curcas] |