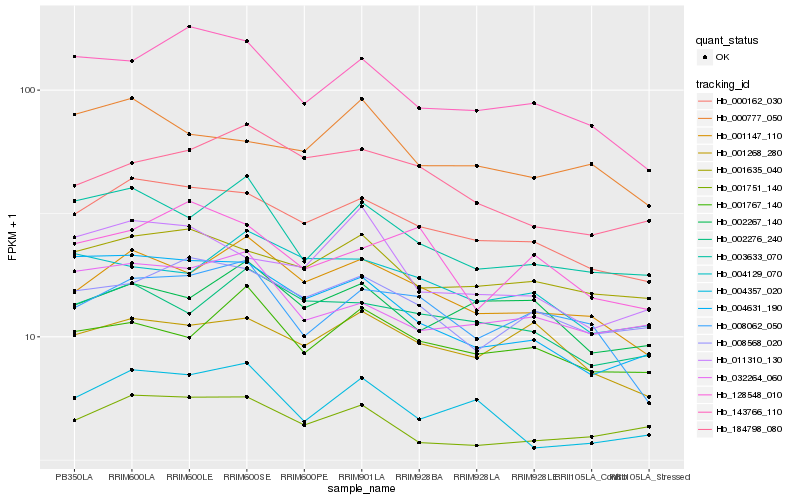
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000162_030 |
0.0 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_001635_040 |
0.0493110898 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 3 |
Hb_001147_110 |
0.0545624809 |
- |
- |
PREDICTED: dynamin-2A-like [Jatropha curcas] |
| 4 |
Hb_001268_280 |
0.0639331467 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_008568_020 |
0.0640433424 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 6 |
Hb_004357_020 |
0.0642532643 |
- |
- |
PREDICTED: uncharacterized protein LOC105637355 [Jatropha curcas] |
| 7 |
Hb_002276_240 |
0.0648750827 |
- |
- |
PREDICTED: uncharacterized protein LOC105640682 [Jatropha curcas] |
| 8 |
Hb_143766_110 |
0.0658389796 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 1 [Jatropha curcas] |
| 9 |
Hb_001751_140 |
0.0727826123 |
- |
- |
PREDICTED: uncharacterized protein LOC105648510 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003633_070 |
0.0734612403 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 11 |
Hb_000777_050 |
0.0738216327 |
- |
- |
hypothetical protein CICLE_v10008422mg [Citrus clementina] |
| 12 |
Hb_002267_140 |
0.0741523469 |
- |
- |
PREDICTED: VHS domain-containing protein At3g16270 [Jatropha curcas] |
| 13 |
Hb_008062_050 |
0.0756619739 |
- |
- |
PREDICTED: uncharacterized protein LOC105631631 [Jatropha curcas] |
| 14 |
Hb_011310_130 |
0.0758335176 |
- |
- |
DEAD/DEAH box RNA helicase family protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_184798_080 |
0.0758755367 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 16 |
Hb_032264_060 |
0.0767237023 |
- |
- |
hypothetical protein JCGZ_02506 [Jatropha curcas] |
| 17 |
Hb_004631_190 |
0.0768312701 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 18 |
Hb_004129_070 |
0.0772585204 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 19 |
Hb_128548_010 |
0.0783217166 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001767_140 |
0.0783756727 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |