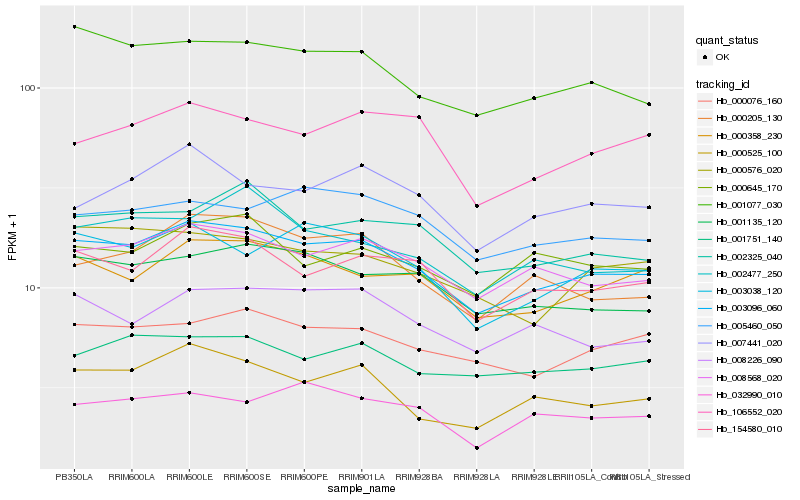
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003096_060 |
0.0 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 2 |
Hb_008568_020 |
0.0513160672 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 3 |
Hb_154580_010 |
0.0563162503 |
- |
- |
hypothetical protein PRUPE_ppa002109mg [Prunus persica] |
| 4 |
Hb_001077_030 |
0.0595313917 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000358_230 |
0.0627926885 |
- |
- |
protein phosphatases pp1 regulatory subunit, putative [Ricinus communis] |
| 6 |
Hb_000076_160 |
0.0642052429 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 7 |
Hb_001135_120 |
0.0642870484 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_003038_120 |
0.0661935966 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 9 |
Hb_002325_040 |
0.0677281771 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 10 |
Hb_008226_090 |
0.0691020506 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000525_100 |
0.0697300292 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 12 |
Hb_005460_050 |
0.0711337857 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 13 |
Hb_001751_140 |
0.0715864067 |
- |
- |
PREDICTED: uncharacterized protein LOC105648510 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000205_130 |
0.072614092 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 15 |
Hb_007441_020 |
0.0739550196 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 16 |
Hb_000576_020 |
0.0741253444 |
- |
- |
PREDICTED: uncharacterized protein LOC105638286 [Jatropha curcas] |
| 17 |
Hb_002477_250 |
0.0747157947 |
- |
- |
DNA-directed RNA polymerase I 49 kDa polypeptide, putative [Ricinus communis] |
| 18 |
Hb_000645_170 |
0.076241136 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_106552_020 |
0.0765862482 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS2Z33 isoform X1 [Jatropha curcas] |
| 20 |
Hb_032990_010 |
0.0778298595 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |