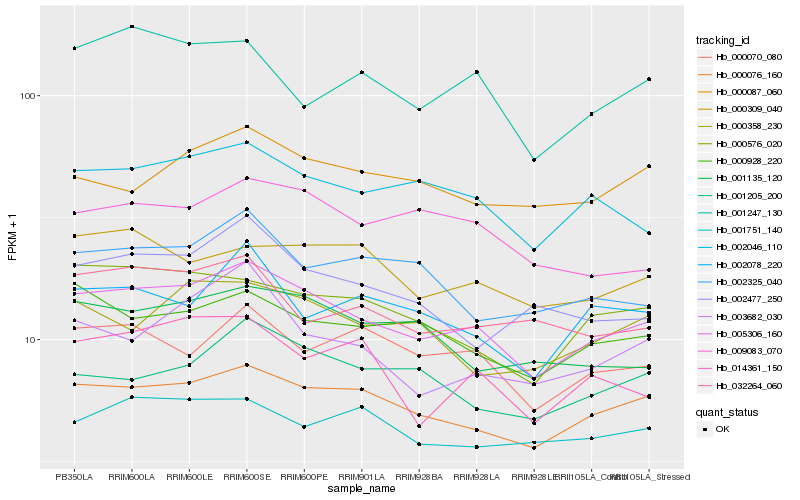
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005306_160 |
0.0 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1-like [Jatropha curcas] |
| 2 |
Hb_000076_160 |
0.0530046307 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 3 |
Hb_002046_110 |
0.0668702375 |
- |
- |
PREDICTED: TBC1 domain family member 15 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000928_220 |
0.0741080995 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 5 |
Hb_000576_020 |
0.0757278046 |
- |
- |
PREDICTED: uncharacterized protein LOC105638286 [Jatropha curcas] |
| 6 |
Hb_000070_080 |
0.0773924765 |
- |
- |
PREDICTED: uncharacterized protein LOC105645742 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000309_040 |
0.078625129 |
- |
- |
PREDICTED: protein farnesyltransferase subunit beta isoform X1 [Jatropha curcas] |
| 8 |
Hb_001205_200 |
0.0792133252 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001135_120 |
0.0793260929 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000358_230 |
0.0811916595 |
- |
- |
protein phosphatases pp1 regulatory subunit, putative [Ricinus communis] |
| 11 |
Hb_009083_070 |
0.0812462553 |
- |
- |
PREDICTED: uncharacterized protein LOC105639870 [Jatropha curcas] |
| 12 |
Hb_002325_040 |
0.0814311781 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000087_060 |
0.0818294656 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 14 |
Hb_003682_030 |
0.0824516902 |
- |
- |
hypothetical protein CICLE_v10020876mg [Citrus clementina] |
| 15 |
Hb_014361_150 |
0.0825031345 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 16 |
Hb_002078_220 |
0.0829370196 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001751_140 |
0.0835649903 |
- |
- |
PREDICTED: uncharacterized protein LOC105648510 isoform X1 [Jatropha curcas] |
| 18 |
Hb_032264_060 |
0.0836352913 |
- |
- |
hypothetical protein JCGZ_02506 [Jatropha curcas] |
| 19 |
Hb_001247_130 |
0.0845767558 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha [Jatropha curcas] |
| 20 |
Hb_002477_250 |
0.0847209972 |
- |
- |
DNA-directed RNA polymerase I 49 kDa polypeptide, putative [Ricinus communis] |