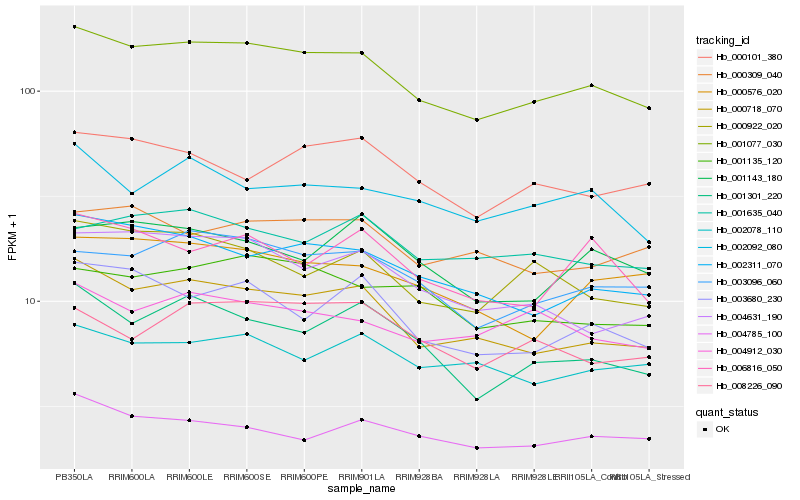
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001077_030 |
0.0 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000718_070 |
0.0510046117 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002311_070 |
0.0593491262 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 4 |
Hb_003096_060 |
0.0595313917 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 5 |
Hb_003680_230 |
0.0715941724 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 6 |
Hb_001135_120 |
0.0718233749 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000922_020 |
0.0734050177 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 8 |
Hb_000576_020 |
0.0734758996 |
- |
- |
PREDICTED: uncharacterized protein LOC105638286 [Jatropha curcas] |
| 9 |
Hb_001301_220 |
0.0738516152 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 4 [Jatropha curcas] |
| 10 |
Hb_004631_190 |
0.0758325412 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 11 |
Hb_002078_110 |
0.0763440112 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 12 |
Hb_002092_080 |
0.0765572856 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform-like [Jatropha curcas] |
| 13 |
Hb_000309_040 |
0.077220615 |
- |
- |
PREDICTED: protein farnesyltransferase subunit beta isoform X1 [Jatropha curcas] |
| 14 |
Hb_004785_100 |
0.0779534501 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000101_380 |
0.0780448652 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 16 |
Hb_004912_030 |
0.0784415809 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 17 |
Hb_001635_040 |
0.0794524457 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 18 |
Hb_006816_050 |
0.0799793527 |
- |
- |
PREDICTED: RNA polymerase II-associated protein 3 isoform X4 [Jatropha curcas] |
| 19 |
Hb_008226_090 |
0.0801278214 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001143_180 |
0.0803712758 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |