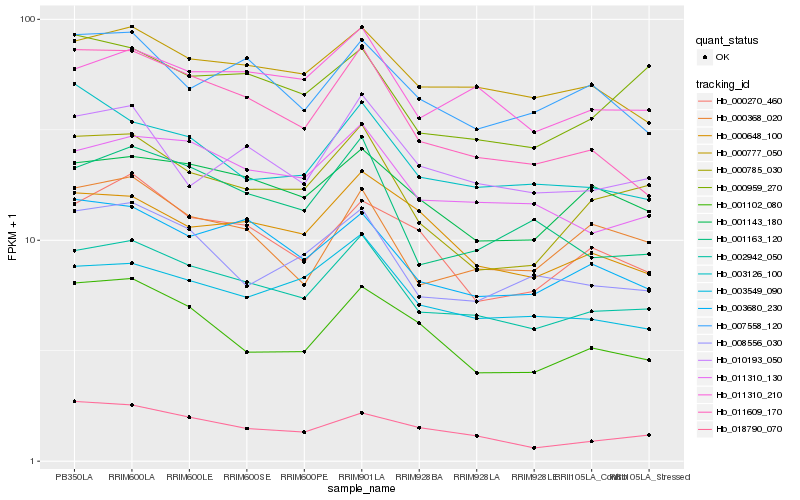
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002942_050 |
0.0 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 2 |
Hb_011310_130 |
0.0588781864 |
- |
- |
DEAD/DEAH box RNA helicase family protein isoform 1 [Theobroma cacao] |
| 3 |
Hb_000777_050 |
0.0644449003 |
- |
- |
hypothetical protein CICLE_v10008422mg [Citrus clementina] |
| 4 |
Hb_003680_230 |
0.0675921672 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 5 |
Hb_011310_210 |
0.0693103591 |
- |
- |
PREDICTED: serine/threonine-protein kinase PRP4 homolog [Jatropha curcas] |
| 6 |
Hb_003549_090 |
0.0704631443 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 6 [Jatropha curcas] |
| 7 |
Hb_001143_180 |
0.0708750207 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |
| 8 |
Hb_008556_030 |
0.0717137392 |
- |
- |
PREDICTED: uncharacterized protein LOC105648021 isoform X1 [Jatropha curcas] |
| 9 |
Hb_011609_170 |
0.0740645982 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 10 |
Hb_001163_120 |
0.0770716952 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 11 |
Hb_000785_030 |
0.0779743568 |
- |
- |
hypothetical protein RCOM_0808030 [Ricinus communis] |
| 12 |
Hb_010193_050 |
0.0800534715 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007558_120 |
0.0806081034 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 14 |
Hb_000648_100 |
0.081662798 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 15 |
Hb_000270_460 |
0.0818583731 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 8 [Jatropha curcas] |
| 16 |
Hb_003126_100 |
0.0821366437 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 17 |
Hb_000368_020 |
0.082639516 |
- |
- |
uroporphyrin-III methyltransferase, putative [Ricinus communis] |
| 18 |
Hb_000959_270 |
0.0839279701 |
- |
- |
PREDICTED: uncharacterized protein LOC105641005 [Jatropha curcas] |
| 19 |
Hb_018790_070 |
0.0842411085 |
- |
- |
PREDICTED: uncharacterized protein LOC105650163 [Jatropha curcas] |
| 20 |
Hb_001102_080 |
0.0844605513 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 4 [Jatropha curcas] |