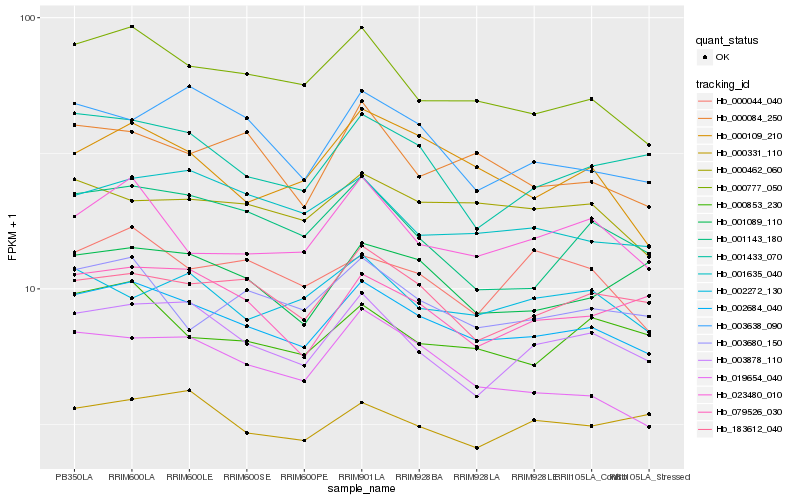
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002684_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000777_050 |
0.0540134409 |
- |
- |
hypothetical protein CICLE_v10008422mg [Citrus clementina] |
| 3 |
Hb_183612_040 |
0.0595959601 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 4 |
Hb_003878_110 |
0.060144028 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 5 |
Hb_000462_060 |
0.0633194412 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001635_040 |
0.0659972453 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 7 |
Hb_000853_230 |
0.066631739 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 8 |
Hb_003680_150 |
0.0667042945 |
- |
- |
srpk, putative [Ricinus communis] |
| 9 |
Hb_023480_010 |
0.066709949 |
- |
- |
cellular nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_079526_030 |
0.0675366458 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 11 |
Hb_019654_040 |
0.0676450549 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase Pus10 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001433_070 |
0.0680080651 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 13 |
Hb_000084_250 |
0.0691226238 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 14 |
Hb_000109_210 |
0.0703412373 |
- |
- |
PREDICTED: threonine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 15 |
Hb_001089_110 |
0.0712213622 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002272_130 |
0.071260822 |
- |
- |
PREDICTED: DNA excision repair protein ERCC-1 [Jatropha curcas] |
| 17 |
Hb_000331_110 |
0.0717304558 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 18 |
Hb_003638_090 |
0.0732508189 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |
| 19 |
Hb_001143_180 |
0.0736054536 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |
| 20 |
Hb_000044_040 |
0.0744637549 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |