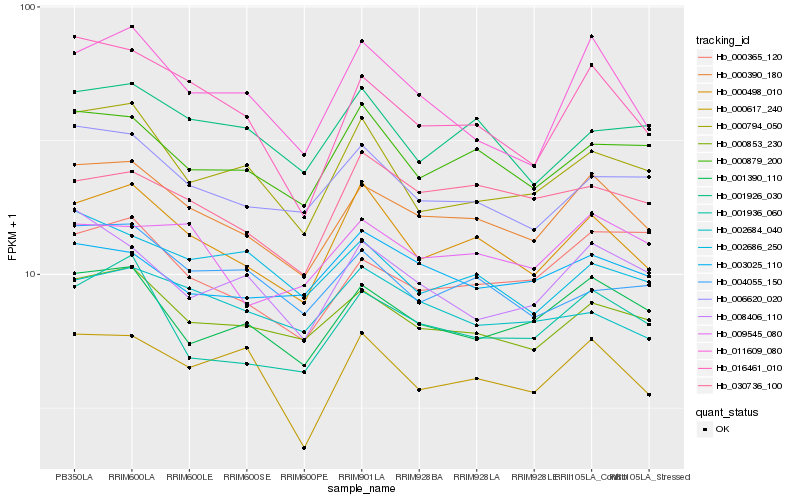
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000390_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC101291673 [Fragaria vesca subsp. vesca] |
| 2 |
Hb_000498_010 |
0.0551125777 |
- |
- |
PREDICTED: protease Do-like 10, mitochondrial isoform X1 [Jatropha curcas] |
| 3 |
Hb_000853_230 |
0.0567817094 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 4 |
Hb_001390_110 |
0.059493866 |
- |
- |
PREDICTED: uncharacterized protein LOC105635984 [Jatropha curcas] |
| 5 |
Hb_016461_010 |
0.0624208184 |
- |
- |
PREDICTED: proline synthase co-transcribed bacterial homolog protein [Jatropha curcas] |
| 6 |
Hb_006620_020 |
0.0670747731 |
- |
- |
PREDICTED: uncharacterized protein LOC105638878 [Jatropha curcas] |
| 7 |
Hb_001936_060 |
0.0678241409 |
- |
- |
pyroglutamyl-peptidase I, putative [Ricinus communis] |
| 8 |
Hb_000879_200 |
0.0688660976 |
- |
- |
PREDICTED: A-kinase anchor protein 17A [Jatropha curcas] |
| 9 |
Hb_009545_080 |
0.073459693 |
- |
- |
hypothetical protein POPTR_0008s15400g [Populus trichocarpa] |
| 10 |
Hb_008406_110 |
0.0759145452 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a isoform X1 [Jatropha curcas] |
| 11 |
Hb_000617_240 |
0.076157363 |
- |
- |
PREDICTED: uncharacterized protein LOC105647502 [Jatropha curcas] |
| 12 |
Hb_011609_080 |
0.0767231227 |
- |
- |
uv excision repair protein rad23, putative [Ricinus communis] |
| 13 |
Hb_000794_050 |
0.0771088569 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 12 [Jatropha curcas] |
| 14 |
Hb_030736_100 |
0.0787744922 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000365_120 |
0.0794532332 |
- |
- |
PREDICTED: bifunctional protein FolD 1, mitochondrial [Jatropha curcas] |
| 16 |
Hb_003025_110 |
0.0808036726 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001926_030 |
0.0809603243 |
- |
- |
PREDICTED: apoptosis inhibitor 5 [Jatropha curcas] |
| 18 |
Hb_002686_250 |
0.081598444 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 65 kDa protein isoform X1 [Jatropha curcas] |
| 19 |
Hb_004055_150 |
0.082029615 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002684_040 |
0.0829208259 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |