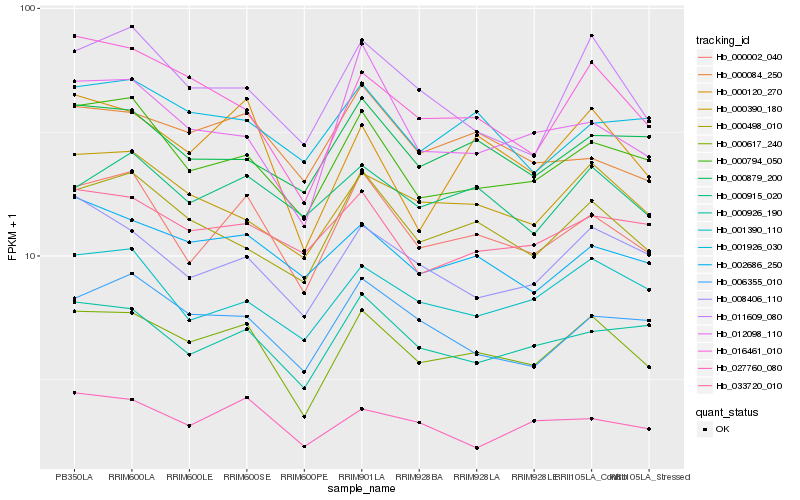
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000617_240 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647502 [Jatropha curcas] |
| 2 |
Hb_000002_040 |
0.0704496293 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 3 |
Hb_016461_010 |
0.0732872541 |
- |
- |
PREDICTED: proline synthase co-transcribed bacterial homolog protein [Jatropha curcas] |
| 4 |
Hb_000390_180 |
0.076157363 |
- |
- |
PREDICTED: uncharacterized protein LOC101291673 [Fragaria vesca subsp. vesca] |
| 5 |
Hb_000794_050 |
0.0780912129 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 12 [Jatropha curcas] |
| 6 |
Hb_000120_270 |
0.0790842908 |
- |
- |
PREDICTED: histone deacetylase 8 [Jatropha curcas] |
| 7 |
Hb_000498_010 |
0.0796189845 |
- |
- |
PREDICTED: protease Do-like 10, mitochondrial isoform X1 [Jatropha curcas] |
| 8 |
Hb_000926_190 |
0.0848615832 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 3, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 9 |
Hb_008406_110 |
0.0862473192 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a isoform X1 [Jatropha curcas] |
| 10 |
Hb_002686_250 |
0.0869579082 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 65 kDa protein isoform X1 [Jatropha curcas] |
| 11 |
Hb_011609_080 |
0.0871294574 |
- |
- |
uv excision repair protein rad23, putative [Ricinus communis] |
| 12 |
Hb_001390_110 |
0.0880873973 |
- |
- |
PREDICTED: uncharacterized protein LOC105635984 [Jatropha curcas] |
| 13 |
Hb_006355_010 |
0.0897858928 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |
| 14 |
Hb_000879_200 |
0.0913544773 |
- |
- |
PREDICTED: A-kinase anchor protein 17A [Jatropha curcas] |
| 15 |
Hb_000915_020 |
0.0925161005 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_000084_250 |
0.0927338793 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 17 |
Hb_033720_010 |
0.0929208594 |
- |
- |
PREDICTED: uncharacterized protein LOC105648843 [Jatropha curcas] |
| 18 |
Hb_027760_080 |
0.0936200245 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_012098_110 |
0.0936436823 |
- |
- |
Ribonuclease 3 [Gossypium arboreum] |
| 20 |
Hb_001926_030 |
0.0940051943 |
- |
- |
PREDICTED: apoptosis inhibitor 5 [Jatropha curcas] |