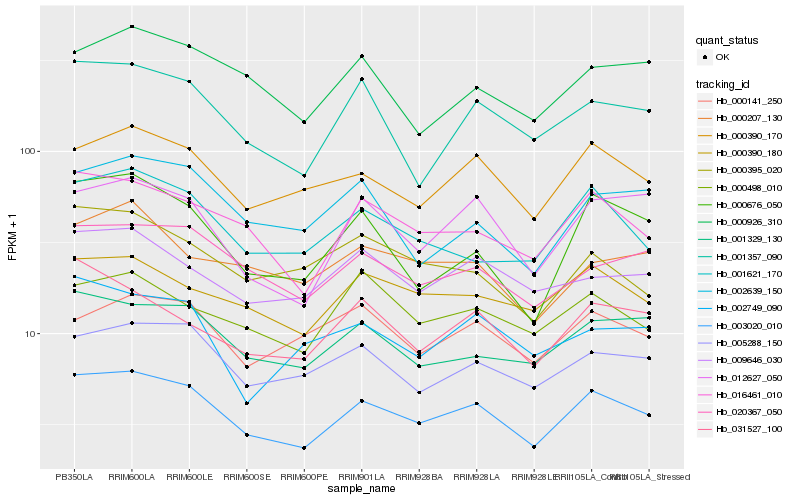
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003020_010 |
0.0 |
transcription factor |
TF Family: G2-like |
PREDICTED: transcription factor LUX [Jatropha curcas] |
| 2 |
Hb_016461_010 |
0.0705937445 |
- |
- |
PREDICTED: proline synthase co-transcribed bacterial homolog protein [Jatropha curcas] |
| 3 |
Hb_000390_170 |
0.0739091664 |
- |
- |
PREDICTED: V-type proton ATPase subunit H [Jatropha curcas] |
| 4 |
Hb_020367_050 |
0.0822925444 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl pyrophosphate synthase, putative [Ricinus communis] |
| 5 |
Hb_005288_150 |
0.0827861116 |
- |
- |
PREDICTED: casein kinase I-like isoform X3 [Jatropha curcas] |
| 6 |
Hb_000390_180 |
0.0839760868 |
- |
- |
PREDICTED: uncharacterized protein LOC101291673 [Fragaria vesca subsp. vesca] |
| 7 |
Hb_001329_130 |
0.0856185442 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_012627_050 |
0.0869918734 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_001621_170 |
0.0874012357 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_001357_090 |
0.0874025059 |
- |
- |
PREDICTED: small ubiquitin-related modifier 1 [Jatropha curcas] |
| 11 |
Hb_009646_030 |
0.0921712157 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_031527_100 |
0.092958082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000676_050 |
0.0929855558 |
- |
- |
PREDICTED: thioredoxin domain-containing protein PLP3B-like [Gossypium raimondii] |
| 14 |
Hb_000498_010 |
0.0964473742 |
- |
- |
PREDICTED: protease Do-like 10, mitochondrial isoform X1 [Jatropha curcas] |
| 15 |
Hb_000926_310 |
0.0991573701 |
- |
- |
uncharacterized protein LOC100500241 [Glycine max] |
| 16 |
Hb_002639_150 |
0.0991706768 |
- |
- |
- |
| 17 |
Hb_000141_250 |
0.1000281913 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 2A isoform X3 [Jatropha curcas] |
| 18 |
Hb_000395_020 |
0.1003611189 |
- |
- |
PREDICTED: ankyrin repeat and KH domain-containing protein R11A8.7 [Jatropha curcas] |
| 19 |
Hb_002749_090 |
0.1006110564 |
- |
- |
- |
| 20 |
Hb_000207_130 |
0.1007915859 |
- |
- |
PREDICTED: calcium-dependent protein kinase 11 [Jatropha curcas] |