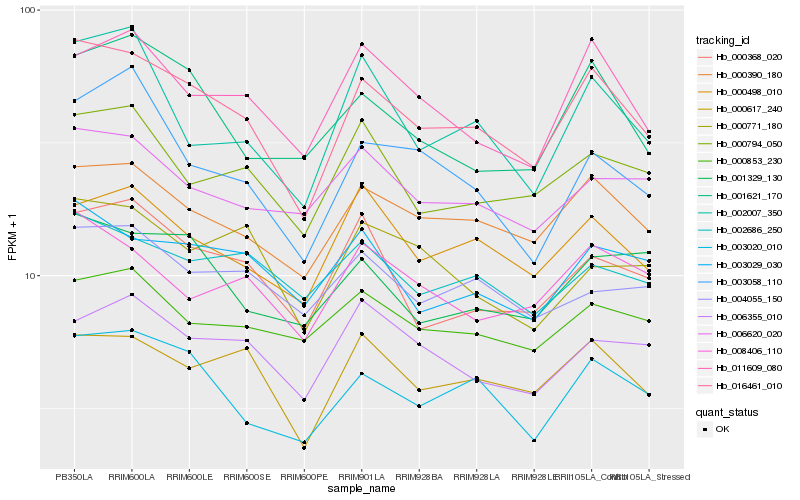
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016461_010 |
0.0 |
- |
- |
PREDICTED: proline synthase co-transcribed bacterial homolog protein [Jatropha curcas] |
| 2 |
Hb_000390_180 |
0.0624208184 |
- |
- |
PREDICTED: uncharacterized protein LOC101291673 [Fragaria vesca subsp. vesca] |
| 3 |
Hb_003020_010 |
0.0705937445 |
transcription factor |
TF Family: G2-like |
PREDICTED: transcription factor LUX [Jatropha curcas] |
| 4 |
Hb_000617_240 |
0.0732872541 |
- |
- |
PREDICTED: uncharacterized protein LOC105647502 [Jatropha curcas] |
| 5 |
Hb_000498_010 |
0.0815715525 |
- |
- |
PREDICTED: protease Do-like 10, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_011609_080 |
0.0816826303 |
- |
- |
uv excision repair protein rad23, putative [Ricinus communis] |
| 7 |
Hb_001621_170 |
0.0874159245 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 8 |
Hb_003029_030 |
0.0877102392 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000794_050 |
0.0879158627 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 12 [Jatropha curcas] |
| 10 |
Hb_000368_020 |
0.0879184969 |
- |
- |
uroporphyrin-III methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_008406_110 |
0.0879471779 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a isoform X1 [Jatropha curcas] |
| 12 |
Hb_002007_350 |
0.0921222836 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 13 |
Hb_002686_250 |
0.0932762866 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 65 kDa protein isoform X1 [Jatropha curcas] |
| 14 |
Hb_001329_130 |
0.0949561666 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006620_020 |
0.0953812687 |
- |
- |
PREDICTED: uncharacterized protein LOC105638878 [Jatropha curcas] |
| 16 |
Hb_003058_110 |
0.0960673253 |
- |
- |
PREDICTED: alpha-mannosidase isoform X2 [Populus euphratica] |
| 17 |
Hb_006355_010 |
0.096709301 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |
| 18 |
Hb_000853_230 |
0.0973221332 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 19 |
Hb_000771_180 |
0.0978345848 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 20 |
Hb_004055_150 |
0.0979114405 |
- |
- |
conserved hypothetical protein [Ricinus communis] |