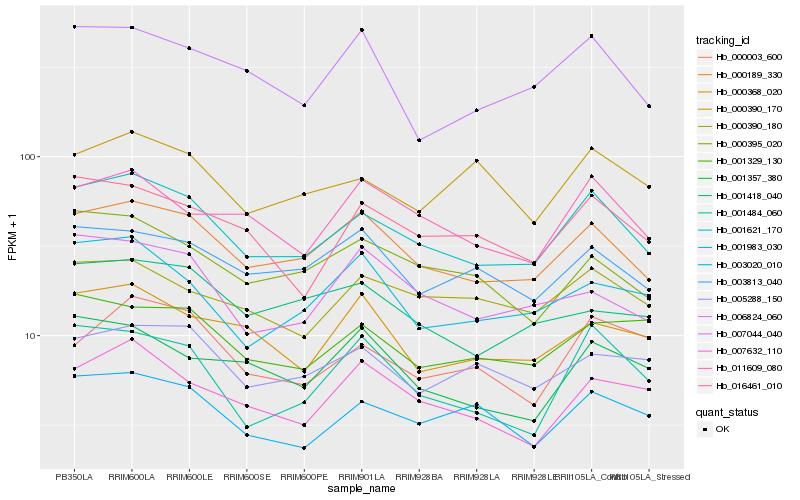
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001621_170 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_000189_330 |
0.0603603126 |
- |
- |
PREDICTED: ER membrane protein complex subunit 2 [Jatropha curcas] |
| 3 |
Hb_003020_010 |
0.0874012357 |
transcription factor |
TF Family: G2-like |
PREDICTED: transcription factor LUX [Jatropha curcas] |
| 4 |
Hb_016461_010 |
0.0874159245 |
- |
- |
PREDICTED: proline synthase co-transcribed bacterial homolog protein [Jatropha curcas] |
| 5 |
Hb_011609_080 |
0.0919260993 |
- |
- |
uv excision repair protein rad23, putative [Ricinus communis] |
| 6 |
Hb_000395_020 |
0.0930647117 |
- |
- |
PREDICTED: ankyrin repeat and KH domain-containing protein R11A8.7 [Jatropha curcas] |
| 7 |
Hb_000390_180 |
0.0947842746 |
- |
- |
PREDICTED: uncharacterized protein LOC101291673 [Fragaria vesca subsp. vesca] |
| 8 |
Hb_003813_040 |
0.0988882496 |
- |
- |
PREDICTED: elongator complex protein 5 [Jatropha curcas] |
| 9 |
Hb_007632_110 |
0.0994148205 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 10 |
Hb_001329_130 |
0.0997879901 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_006824_060 |
0.1001979258 |
- |
- |
PREDICTED: uncharacterized protein LOC105650737 [Jatropha curcas] |
| 12 |
Hb_001484_060 |
0.1004270387 |
- |
- |
PREDICTED: uncharacterized protein LOC105641630 [Jatropha curcas] |
| 13 |
Hb_005288_150 |
0.1012209572 |
- |
- |
PREDICTED: casein kinase I-like isoform X3 [Jatropha curcas] |
| 14 |
Hb_001983_030 |
0.1027776721 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 12 isoform X2 [Jatropha curcas] |
| 15 |
Hb_007044_040 |
0.1044546983 |
- |
- |
PREDICTED: uncharacterized protein LOC105646814 [Jatropha curcas] |
| 16 |
Hb_000368_020 |
0.1045714864 |
- |
- |
uroporphyrin-III methyltransferase, putative [Ricinus communis] |
| 17 |
Hb_000390_170 |
0.1046704544 |
- |
- |
PREDICTED: V-type proton ATPase subunit H [Jatropha curcas] |
| 18 |
Hb_000003_600 |
0.1052494514 |
- |
- |
PREDICTED: protein SAWADEE HOMEODOMAIN HOMOLOG 1 [Jatropha curcas] |
| 19 |
Hb_001357_380 |
0.1056945467 |
- |
- |
PREDICTED: probable tetraacyldisaccharide 4'-kinase, mitochondrial isoform X1 [Jatropha curcas] |
| 20 |
Hb_001418_040 |
0.1058290116 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1-like [Jatropha curcas] |