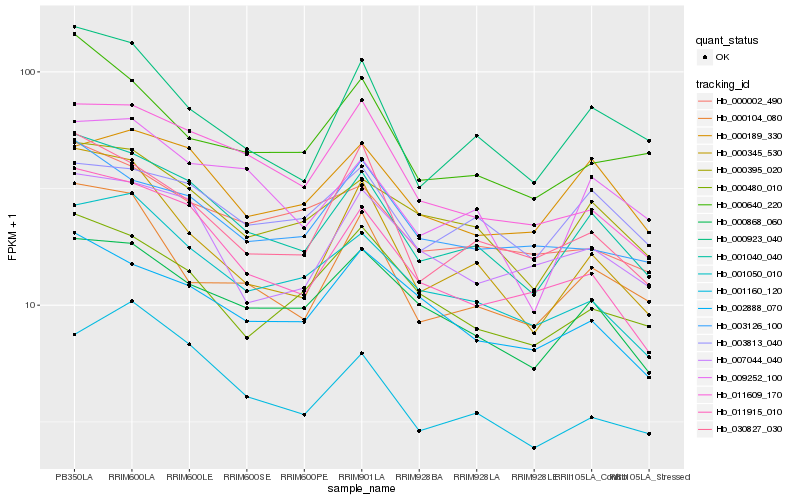
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001040_040 |
0.0 |
- |
- |
Uncharacterized protein TCM_029905 [Theobroma cacao] |
| 2 |
Hb_000395_020 |
0.0702845545 |
- |
- |
PREDICTED: ankyrin repeat and KH domain-containing protein R11A8.7 [Jatropha curcas] |
| 3 |
Hb_011915_010 |
0.072354767 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 4 |
Hb_000868_060 |
0.0733270905 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 5 |
Hb_000923_040 |
0.073825853 |
- |
- |
PREDICTED: 15 kDa selenoprotein [Jatropha curcas] |
| 6 |
Hb_009252_100 |
0.0768394409 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 7 |
Hb_030827_030 |
0.0779431419 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase XBAT33 [Jatropha curcas] |
| 8 |
Hb_000345_530 |
0.0780201445 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001050_010 |
0.0803791007 |
- |
- |
Advillin [Gossypium arboreum] |
| 10 |
Hb_000104_080 |
0.0828392526 |
- |
- |
PREDICTED: WD repeat domain-containing protein 83 [Jatropha curcas] |
| 11 |
Hb_000002_490 |
0.0881623038 |
- |
- |
PREDICTED: uncharacterized protein LOC105632095 [Jatropha curcas] |
| 12 |
Hb_003126_100 |
0.089749431 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 13 |
Hb_002888_070 |
0.0903674109 |
- |
- |
PREDICTED: tubulin-folding cofactor E isoform X1 [Pyrus x bretschneideri] |
| 14 |
Hb_011609_170 |
0.091244857 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 15 |
Hb_000640_220 |
0.0916389488 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-4 [Jatropha curcas] |
| 16 |
Hb_007044_040 |
0.0924039827 |
- |
- |
PREDICTED: uncharacterized protein LOC105646814 [Jatropha curcas] |
| 17 |
Hb_003813_040 |
0.0926358613 |
- |
- |
PREDICTED: elongator complex protein 5 [Jatropha curcas] |
| 18 |
Hb_000480_010 |
0.0926854932 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001160_120 |
0.0938584612 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000189_330 |
0.0944329149 |
- |
- |
PREDICTED: ER membrane protein complex subunit 2 [Jatropha curcas] |