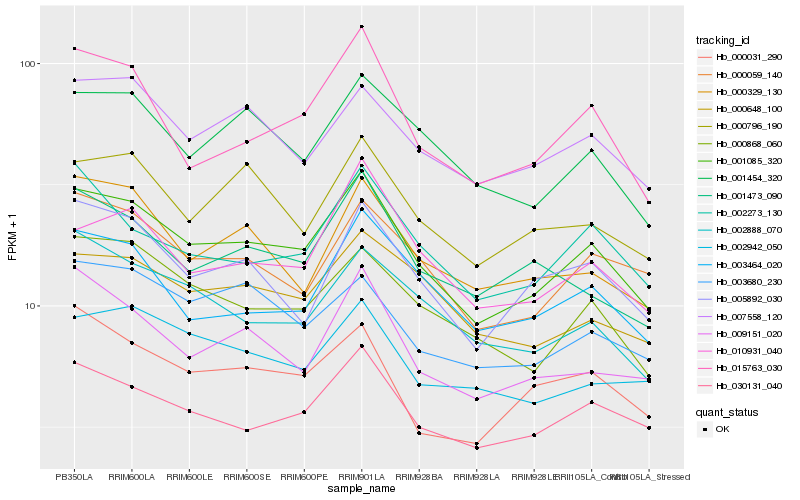
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001085_320 |
0.0 |
transcription factor |
TF Family: LUG |
WD-repeat protein, putative [Ricinus communis] |
| 2 |
Hb_030131_040 |
0.066880468 |
- |
- |
ribosome biogenesis protein tsr1, putative [Ricinus communis] |
| 3 |
Hb_000868_060 |
0.0720596569 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 4 |
Hb_000059_140 |
0.0747633678 |
- |
- |
rnase h, putative [Ricinus communis] |
| 5 |
Hb_015763_030 |
0.0758370503 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007558_120 |
0.0758691967 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 7 |
Hb_001454_320 |
0.0771950365 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_005892_030 |
0.0780359106 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 9 |
Hb_002273_130 |
0.0780414281 |
- |
- |
PREDICTED: T-complex protein 1 subunit eta [Gossypium raimondii] |
| 10 |
Hb_000648_100 |
0.0782008108 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 11 |
Hb_010931_040 |
0.0795331598 |
- |
- |
PREDICTED: nijmegen breakage syndrome 1 protein [Jatropha curcas] |
| 12 |
Hb_000796_190 |
0.079948552 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 13 |
Hb_009151_020 |
0.0804966077 |
- |
- |
PREDICTED: DNA polymerase delta catalytic subunit [Jatropha curcas] |
| 14 |
Hb_003464_020 |
0.0809518517 |
- |
- |
srpk, putative [Ricinus communis] |
| 15 |
Hb_002888_070 |
0.0813571503 |
- |
- |
PREDICTED: tubulin-folding cofactor E isoform X1 [Pyrus x bretschneideri] |
| 16 |
Hb_003680_230 |
0.082594453 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 17 |
Hb_000329_130 |
0.082638683 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 18 |
Hb_000031_290 |
0.0829995705 |
- |
- |
hypothetical protein POPTR_0006s28540g [Populus trichocarpa] |
| 19 |
Hb_001473_090 |
0.0839884615 |
- |
- |
PREDICTED: uncharacterized protein LOC105650374 [Jatropha curcas] |
| 20 |
Hb_002942_050 |
0.0845357172 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |