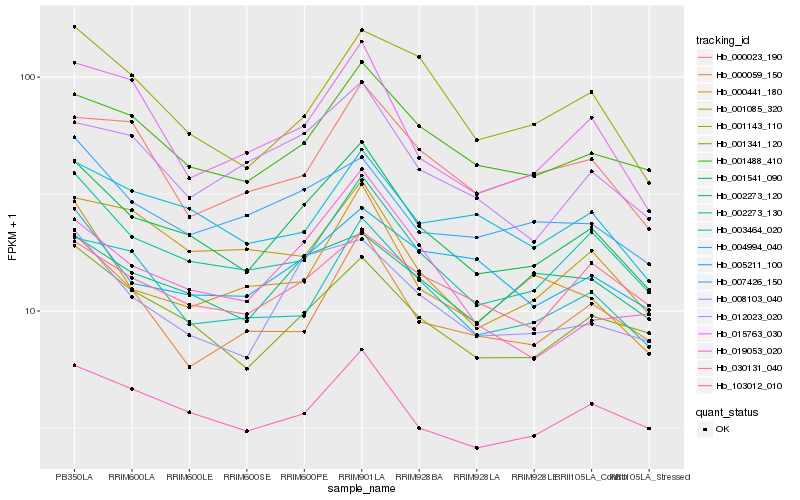
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001541_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002273_130 |
0.0666567734 |
- |
- |
PREDICTED: T-complex protein 1 subunit eta [Gossypium raimondii] |
| 3 |
Hb_001341_120 |
0.0739623591 |
- |
- |
hypothetical protein B456_001G130300 [Gossypium raimondii] |
| 4 |
Hb_030131_040 |
0.0742213907 |
- |
- |
ribosome biogenesis protein tsr1, putative [Ricinus communis] |
| 5 |
Hb_008103_040 |
0.074411213 |
- |
- |
diacylglycerol O-acyltransferase 1 [Jatropha curcas] |
| 6 |
Hb_001488_410 |
0.0745689486 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 7 |
Hb_003464_020 |
0.0877249021 |
- |
- |
srpk, putative [Ricinus communis] |
| 8 |
Hb_012023_020 |
0.0880452778 |
- |
- |
hypothetical protein JCGZ_03076 [Jatropha curcas] |
| 9 |
Hb_015763_030 |
0.0895604507 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007426_150 |
0.0905885495 |
- |
- |
PREDICTED: F-box protein At5g46170-like [Jatropha curcas] |
| 11 |
Hb_004994_040 |
0.0916104548 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 12 |
Hb_000441_180 |
0.0920954429 |
- |
- |
Early endosome antigen, putative [Ricinus communis] |
| 13 |
Hb_002273_120 |
0.0924968171 |
- |
- |
PREDICTED: probable apyrase 6 [Jatropha curcas] |
| 14 |
Hb_001085_320 |
0.0947404981 |
transcription factor |
TF Family: LUG |
WD-repeat protein, putative [Ricinus communis] |
| 15 |
Hb_001143_110 |
0.0950799838 |
- |
- |
hypothetical protein JCGZ_12107 [Jatropha curcas] |
| 16 |
Hb_103012_010 |
0.0951845937 |
- |
- |
PREDICTED: uncharacterized protein LOC105638878 [Jatropha curcas] |
| 17 |
Hb_005211_100 |
0.0953445897 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform [Jatropha curcas] |
| 18 |
Hb_019053_020 |
0.0972642417 |
- |
- |
hypothetical protein MIMGU_mgv1a0105441mg, partial [Erythranthe guttata] |
| 19 |
Hb_000023_190 |
0.0975631833 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 20 |
Hb_000059_150 |
0.0983169666 |
- |
- |
Polynucleotidyl transferase, ribonuclease H-like superfamily protein isoform 2 [Theobroma cacao] |