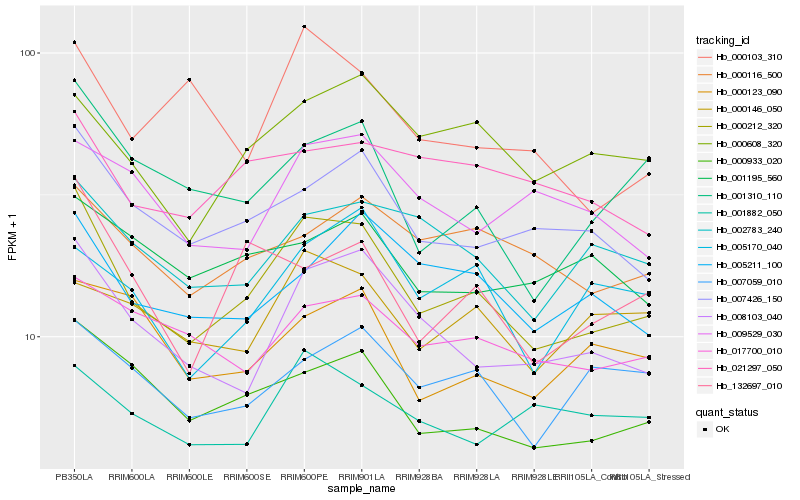
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000212_320 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644891 [Jatropha curcas] |
| 2 |
Hb_008103_040 |
0.0919347604 |
- |
- |
diacylglycerol O-acyltransferase 1 [Jatropha curcas] |
| 3 |
Hb_000933_020 |
0.0946680172 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_007426_150 |
0.0983133982 |
- |
- |
PREDICTED: F-box protein At5g46170-like [Jatropha curcas] |
| 5 |
Hb_000608_320 |
0.1086146843 |
- |
- |
PREDICTED: uncharacterized protein LOC105644891 [Jatropha curcas] |
| 6 |
Hb_132697_010 |
0.1096794434 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001310_110 |
0.1103011122 |
- |
- |
unnamed protein product [Coffea canephora] |
| 8 |
Hb_002783_240 |
0.1120416422 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 9 |
Hb_005211_100 |
0.1122445743 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform [Jatropha curcas] |
| 10 |
Hb_007059_010 |
0.1164405027 |
- |
- |
PREDICTED: uncharacterized protein LOC105642418 [Jatropha curcas] |
| 11 |
Hb_000116_500 |
0.1169005058 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 11 [Jatropha curcas] |
| 12 |
Hb_017700_010 |
0.1177202434 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 13 |
Hb_005170_040 |
0.1205608953 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Jatropha curcas] |
| 14 |
Hb_000146_050 |
0.1214095822 |
- |
- |
PREDICTED: protein unc-50 homolog isoform X2 [Gossypium raimondii] |
| 15 |
Hb_021297_050 |
0.1232231787 |
- |
- |
PREDICTED: signal recognition particle receptor subunit alpha [Jatropha curcas] |
| 16 |
Hb_000123_090 |
0.1235092416 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os03g0620400-like [Jatropha curcas] |
| 17 |
Hb_009529_030 |
0.1250357327 |
- |
- |
PREDICTED: transmembrane protein 120 homolog [Jatropha curcas] |
| 18 |
Hb_001882_050 |
0.1255326011 |
- |
- |
hydroxysteroid dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_001195_560 |
0.1258413436 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000103_310 |
0.127119937 |
- |
- |
PREDICTED: transmembrane emp24 domain-containing protein p24beta3 [Jatropha curcas] |