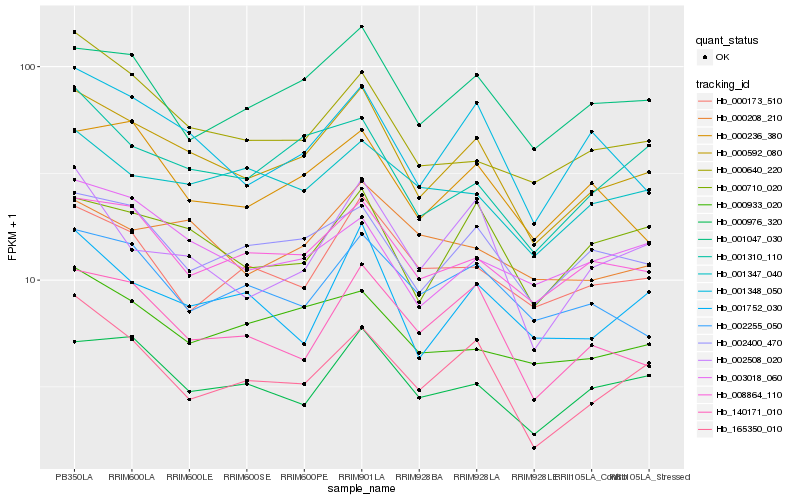
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000592_080 |
0.0 |
- |
- |
PREDICTED: thymidylate kinase [Jatropha curcas] |
| 2 |
Hb_008864_110 |
0.0845342668 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 3 |
Hb_002400_470 |
0.0871870872 |
- |
- |
PREDICTED: calcium uptake protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 4 |
Hb_001348_050 |
0.0875562642 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001047_030 |
0.0885201974 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 6 |
Hb_000976_320 |
0.0928033193 |
- |
- |
UPF0061 protein azo1574 [Morus notabilis] |
| 7 |
Hb_000710_020 |
0.0935925562 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MARCH2 isoform X2 [Populus euphratica] |
| 8 |
Hb_140171_010 |
0.0959829793 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |
| 9 |
Hb_001310_110 |
0.0981433369 |
- |
- |
unnamed protein product [Coffea canephora] |
| 10 |
Hb_000236_380 |
0.100210537 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PK [Jatropha curcas] |
| 11 |
Hb_001752_030 |
0.1018209846 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 4 [Jatropha curcas] |
| 12 |
Hb_165350_010 |
0.101935293 |
- |
- |
unnamed protein product [Triticum aestivum] |
| 13 |
Hb_002508_020 |
0.1031638354 |
- |
- |
PREDICTED: SNF1-related protein kinase regulatory subunit beta-1 [Jatropha curcas] |
| 14 |
Hb_002255_050 |
0.104118849 |
- |
- |
PREDICTED: probable isoleucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 15 |
Hb_000640_220 |
0.1043646883 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-4 [Jatropha curcas] |
| 16 |
Hb_000173_510 |
0.1046878383 |
- |
- |
PREDICTED: AP-5 complex subunit mu [Jatropha curcas] |
| 17 |
Hb_001347_040 |
0.1048511407 |
- |
- |
PREDICTED: uncharacterized protein LOC105633920 [Jatropha curcas] |
| 18 |
Hb_000933_020 |
0.1050381682 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000208_210 |
0.1050627105 |
- |
- |
PREDICTED: RNA-binding protein Nova-2 [Jatropha curcas] |
| 20 |
Hb_003018_060 |
0.1064336099 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase A5 [Jatropha curcas] |