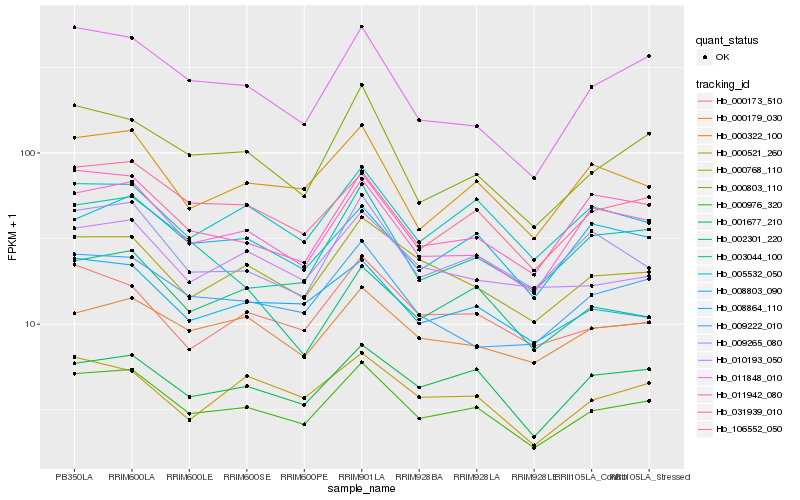
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000976_320 |
0.0 |
- |
- |
UPF0061 protein azo1574 [Morus notabilis] |
| 2 |
Hb_106552_050 |
0.0631374649 |
- |
- |
PREDICTED: uncharacterized protein LOC105642134 [Jatropha curcas] |
| 3 |
Hb_011942_080 |
0.0669345002 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 4 |
Hb_008864_110 |
0.0701849779 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 5 |
Hb_000768_110 |
0.0710700637 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000322_100 |
0.0722601072 |
- |
- |
PREDICTED: casein kinase I isoform delta-like [Jatropha curcas] |
| 7 |
Hb_001677_210 |
0.0735038538 |
- |
- |
hypothetical protein JCGZ_01374 [Jatropha curcas] |
| 8 |
Hb_005532_050 |
0.0757992484 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 9 |
Hb_000803_110 |
0.0769906925 |
- |
- |
PREDICTED: RNA-binding protein 8A [Jatropha curcas] |
| 10 |
Hb_008803_090 |
0.0798806739 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 6a [Jatropha curcas] |
| 11 |
Hb_031939_010 |
0.0807936052 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 12 |
Hb_000521_260 |
0.0818255527 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_010193_050 |
0.0824585288 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_009265_080 |
0.0827080014 |
- |
- |
PREDICTED: apoptosis inhibitor 5 [Jatropha curcas] |
| 15 |
Hb_000179_030 |
0.0832569673 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002301_220 |
0.0833492402 |
transcription factor |
TF Family: C3H |
PREDICTED: protein FRIGIDA-ESSENTIAL 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000173_510 |
0.0833563304 |
- |
- |
PREDICTED: AP-5 complex subunit mu [Jatropha curcas] |
| 18 |
Hb_009222_010 |
0.0835726429 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 19 |
Hb_003044_100 |
0.0845924715 |
- |
- |
hypothetical protein F383_08446 [Gossypium arboreum] |
| 20 |
Hb_011848_010 |
0.0860868895 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |