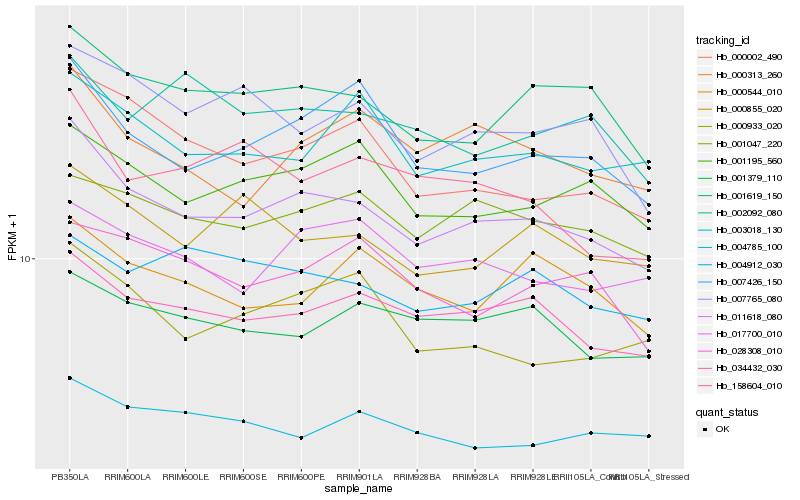
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011618_080 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 2 |
Hb_034432_030 |
0.0702611141 |
- |
- |
PREDICTED: probable glycosyltransferase At5g20260 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001619_150 |
0.0738288968 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000002_490 |
0.0780663019 |
- |
- |
PREDICTED: uncharacterized protein LOC105632095 [Jatropha curcas] |
| 5 |
Hb_007426_150 |
0.0793793429 |
- |
- |
PREDICTED: F-box protein At5g46170-like [Jatropha curcas] |
| 6 |
Hb_001195_560 |
0.0847422845 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_007765_080 |
0.0853818845 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000855_020 |
0.0864425282 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Populus euphratica] |
| 9 |
Hb_000313_260 |
0.0874818233 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 10 |
Hb_003018_130 |
0.0881803778 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 11 |
Hb_000933_020 |
0.0901808641 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_017700_010 |
0.0902325486 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 13 |
Hb_001047_220 |
0.0906358123 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 14 |
Hb_001379_110 |
0.0921628803 |
- |
- |
PREDICTED: sec1 family domain-containing protein MIP3 [Jatropha curcas] |
| 15 |
Hb_158604_010 |
0.0939856523 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 16 |
Hb_004785_100 |
0.0948526178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000544_010 |
0.0975517536 |
- |
- |
PREDICTED: GPI mannosyltransferase 1 [Jatropha curcas] |
| 18 |
Hb_028308_010 |
0.0975923063 |
- |
- |
PREDICTED: probable protein S-acyltransferase 4 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004912_030 |
0.0987075235 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 20 |
Hb_002092_080 |
0.099247408 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform-like [Jatropha curcas] |