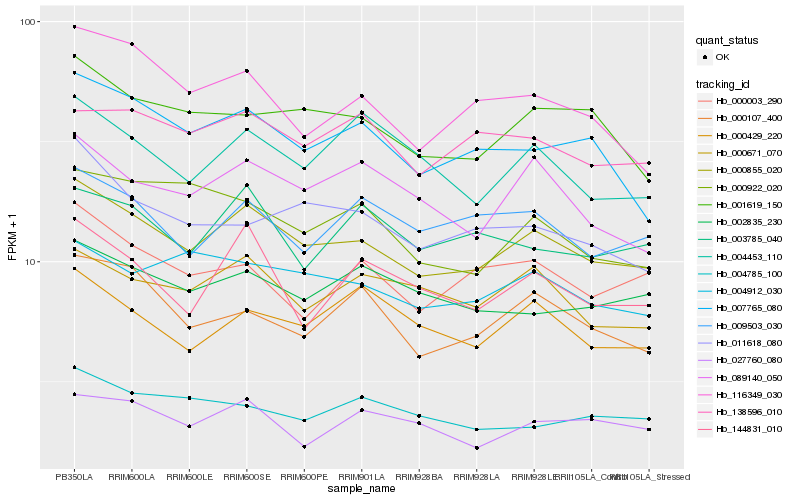
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000855_020 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Populus euphratica] |
| 2 |
Hb_001619_150 |
0.0746821629 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_007765_080 |
0.0769030962 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000671_070 |
0.0773451757 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 5 |
Hb_004912_030 |
0.078785884 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 6 |
Hb_089140_050 |
0.0788020623 |
- |
- |
hypothetical protein JCGZ_07485 [Jatropha curcas] |
| 7 |
Hb_002835_230 |
0.0818087903 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_009503_030 |
0.0825695637 |
- |
- |
protein dimerization, putative [Ricinus communis] |
| 9 |
Hb_000107_400 |
0.0826529611 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46870-like [Jatropha curcas] |
| 10 |
Hb_116349_030 |
0.0833562835 |
- |
- |
PREDICTED: zinc finger MYND domain-containing protein 15 isoform X3 [Jatropha curcas] |
| 11 |
Hb_138596_010 |
0.0842147614 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g13690 [Jatropha curcas] |
| 12 |
Hb_000429_220 |
0.0850513866 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105640858 isoform X1 [Jatropha curcas] |
| 13 |
Hb_144831_010 |
0.0852473814 |
transcription factor |
TF Family: MYB |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_004453_110 |
0.0858781532 |
- |
- |
Peptidyl-prolyl cis-trans isomerase CYP19-2 isoform 1 [Theobroma cacao] |
| 15 |
Hb_011618_080 |
0.0864425282 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 16 |
Hb_003785_040 |
0.086843562 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000003_290 |
0.0869311766 |
- |
- |
hypothetical protein JCGZ_24486 [Jatropha curcas] |
| 18 |
Hb_027760_080 |
0.0876017938 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_000922_020 |
0.0887186591 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_004785_100 |
0.0890326144 |
- |
- |
conserved hypothetical protein [Ricinus communis] |