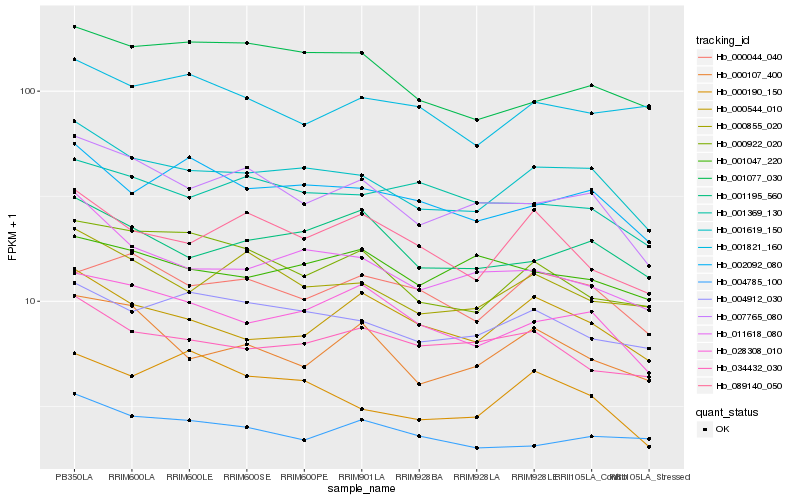
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001619_150 |
0.0 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007765_080 |
0.0720017934 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002092_080 |
0.0720464035 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform-like [Jatropha curcas] |
| 4 |
Hb_028308_010 |
0.0723769006 |
- |
- |
PREDICTED: probable protein S-acyltransferase 4 isoform X1 [Jatropha curcas] |
| 5 |
Hb_011618_080 |
0.0738288968 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 6 |
Hb_000855_020 |
0.0746821629 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Populus euphratica] |
| 7 |
Hb_004912_030 |
0.0749250066 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 8 |
Hb_001195_560 |
0.0771165642 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000544_010 |
0.082053523 |
- |
- |
PREDICTED: GPI mannosyltransferase 1 [Jatropha curcas] |
| 10 |
Hb_034432_030 |
0.0880922136 |
- |
- |
PREDICTED: probable glycosyltransferase At5g20260 isoform X1 [Jatropha curcas] |
| 11 |
Hb_089140_050 |
0.0902071376 |
- |
- |
hypothetical protein JCGZ_07485 [Jatropha curcas] |
| 12 |
Hb_001077_030 |
0.0904203498 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004785_100 |
0.0907298078 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000922_020 |
0.0915233399 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_000107_400 |
0.0918428054 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46870-like [Jatropha curcas] |
| 16 |
Hb_001369_130 |
0.0922315714 |
- |
- |
PREDICTED: autophagy-related protein 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000044_040 |
0.0927388645 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 18 |
Hb_001047_220 |
0.0952611639 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 19 |
Hb_000190_150 |
0.0963469265 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 4B-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_001821_160 |
0.0974984437 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |