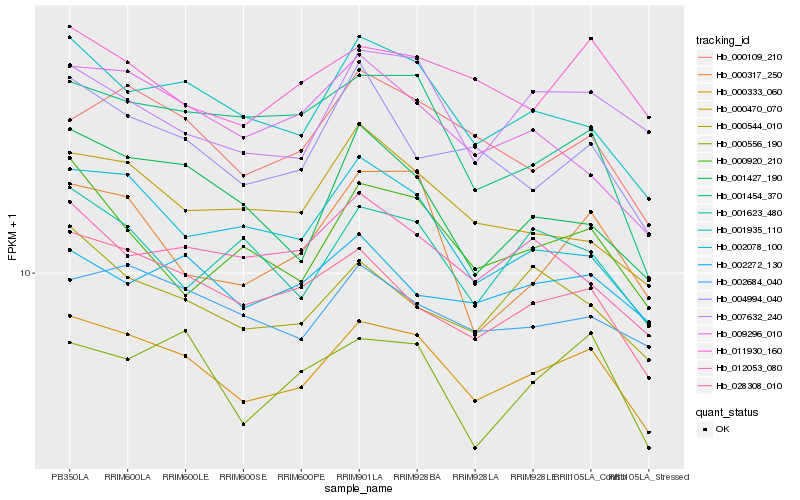
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000333_060 |
0.0 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 2 |
Hb_001935_110 |
0.0626034095 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |
| 3 |
Hb_028308_010 |
0.0711364434 |
- |
- |
PREDICTED: probable protein S-acyltransferase 4 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000109_210 |
0.0733535715 |
- |
- |
PREDICTED: threonine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 5 |
Hb_002684_040 |
0.0750392964 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002078_100 |
0.0780699616 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 7 |
Hb_007632_240 |
0.0795061514 |
- |
- |
transporter, putative [Ricinus communis] |
| 8 |
Hb_000544_010 |
0.0797330659 |
- |
- |
PREDICTED: GPI mannosyltransferase 1 [Jatropha curcas] |
| 9 |
Hb_000556_190 |
0.0820286727 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_011930_160 |
0.0821865411 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 38-like [Jatropha curcas] |
| 11 |
Hb_001427_190 |
0.0832551769 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 12 |
Hb_004994_040 |
0.0843570027 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 13 |
Hb_000470_070 |
0.0843687782 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |
| 14 |
Hb_002272_130 |
0.0868397616 |
- |
- |
PREDICTED: DNA excision repair protein ERCC-1 [Jatropha curcas] |
| 15 |
Hb_009296_010 |
0.0872201748 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 16 |
Hb_012053_080 |
0.0873129555 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 17 |
Hb_001623_480 |
0.0873571307 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 13 [Jatropha curcas] |
| 18 |
Hb_000920_210 |
0.08760156 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 19 |
Hb_001454_370 |
0.0876470637 |
- |
- |
PREDICTED: glyoxysomal fatty acid beta-oxidation multifunctional protein MFP-a [Jatropha curcas] |
| 20 |
Hb_000317_250 |
0.087684934 |
- |
- |
PREDICTED: calcium-dependent protein kinase SK5 isoform X1 [Jatropha curcas] |