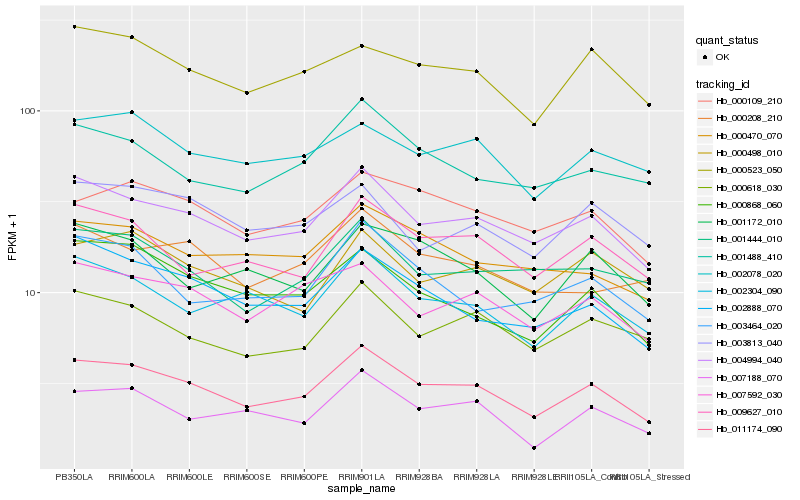
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011174_090 |
0.0 |
- |
- |
PREDICTED: fidgetin-like protein 1 [Jatropha curcas] |
| 2 |
Hb_004994_040 |
0.0448082636 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 3 |
Hb_007592_030 |
0.0757390717 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 4 |
Hb_002304_090 |
0.0768399385 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX35 isoform X1 [Jatropha curcas] |
| 5 |
Hb_009627_010 |
0.0773720442 |
- |
- |
Protein dom-3, putative [Ricinus communis] |
| 6 |
Hb_007188_070 |
0.0776630566 |
- |
- |
prokaryotic DNA topoisomerase, putative [Ricinus communis] |
| 7 |
Hb_000109_210 |
0.078886842 |
- |
- |
PREDICTED: threonine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000523_050 |
0.0809984038 |
- |
- |
microsomal glutathione s-transferase, putative [Ricinus communis] |
| 9 |
Hb_000618_030 |
0.0818030557 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOUSLED-like isoform X4 [Populus euphratica] |
| 10 |
Hb_000868_060 |
0.0827674292 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 11 |
Hb_002888_070 |
0.0830234257 |
- |
- |
PREDICTED: tubulin-folding cofactor E isoform X1 [Pyrus x bretschneideri] |
| 12 |
Hb_001488_410 |
0.0846006059 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 13 |
Hb_000470_070 |
0.0846622055 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |
| 14 |
Hb_002078_020 |
0.0864440182 |
- |
- |
PREDICTED: proteasome subunit beta type-5 [Jatropha curcas] |
| 15 |
Hb_001172_010 |
0.0867996062 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 16 |
Hb_003464_020 |
0.0871562544 |
- |
- |
srpk, putative [Ricinus communis] |
| 17 |
Hb_000498_010 |
0.0903713574 |
- |
- |
PREDICTED: protease Do-like 10, mitochondrial isoform X1 [Jatropha curcas] |
| 18 |
Hb_001444_010 |
0.0904037402 |
transcription factor |
TF Family: DDT |
PREDICTED: DDT domain-containing protein DDB_G0282237 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000208_210 |
0.0908211234 |
- |
- |
PREDICTED: RNA-binding protein Nova-2 [Jatropha curcas] |
| 20 |
Hb_003813_040 |
0.0914866202 |
- |
- |
PREDICTED: elongator complex protein 5 [Jatropha curcas] |