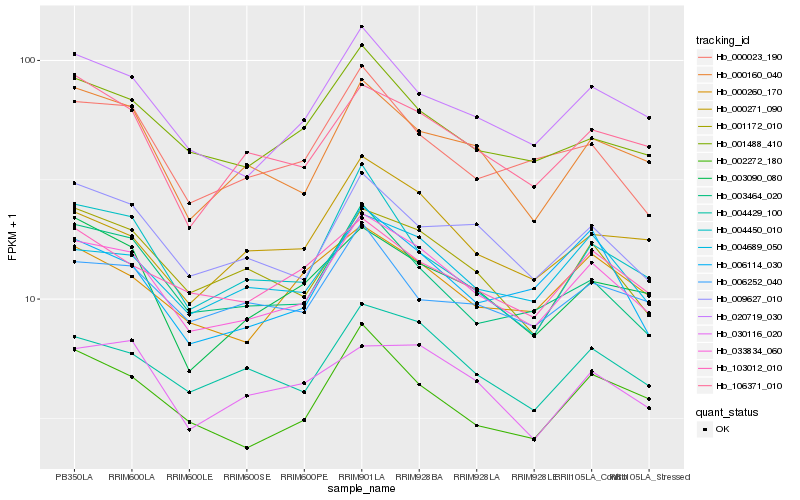
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033834_060 |
0.0 |
- |
- |
DNA-binding bromodomain-containing family protein [Populus trichocarpa] |
| 2 |
Hb_020719_030 |
0.0550505408 |
- |
- |
PREDICTED: ER membrane protein complex subunit 3-like [Jatropha curcas] |
| 3 |
Hb_004689_050 |
0.0580862539 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004429_100 |
0.0591720971 |
- |
- |
PREDICTED: asparagine synthetase domain-containing protein 1 [Jatropha curcas] |
| 5 |
Hb_009627_010 |
0.0641574335 |
- |
- |
Protein dom-3, putative [Ricinus communis] |
| 6 |
Hb_001172_010 |
0.0666679936 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 7 |
Hb_001488_410 |
0.0674656908 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 8 |
Hb_000160_040 |
0.0679867813 |
- |
- |
PREDICTED: methyltransferase-like protein 10 [Jatropha curcas] |
| 9 |
Hb_106371_010 |
0.0706295633 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 10 |
Hb_103012_010 |
0.0724160302 |
- |
- |
PREDICTED: uncharacterized protein LOC105638878 [Jatropha curcas] |
| 11 |
Hb_000023_190 |
0.0728478579 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 12 |
Hb_000260_170 |
0.0736560295 |
- |
- |
PREDICTED: putative zinc transporter At3g08650 [Cucumis sativus] |
| 13 |
Hb_030116_020 |
0.0739741551 |
- |
- |
hypothetical protein POPTR_0016s13630g [Populus trichocarpa] |
| 14 |
Hb_003464_020 |
0.0743089999 |
- |
- |
srpk, putative [Ricinus communis] |
| 15 |
Hb_000271_090 |
0.0758108481 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 16 |
Hb_002272_180 |
0.0762513689 |
- |
- |
hypothetical protein JCGZ_23380 [Jatropha curcas] |
| 17 |
Hb_006114_030 |
0.0768594629 |
transcription factor |
TF Family: SET |
hypothetical protein CISIN_1g011639mg [Citrus sinensis] |
| 18 |
Hb_003090_080 |
0.077506084 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-like protein B [Jatropha curcas] |
| 19 |
Hb_006252_040 |
0.0782450557 |
- |
- |
queuine tRNA-ribosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_004450_010 |
0.0787190684 |
- |
- |
PREDICTED: methyltransferase-like protein 10 [Jatropha curcas] |