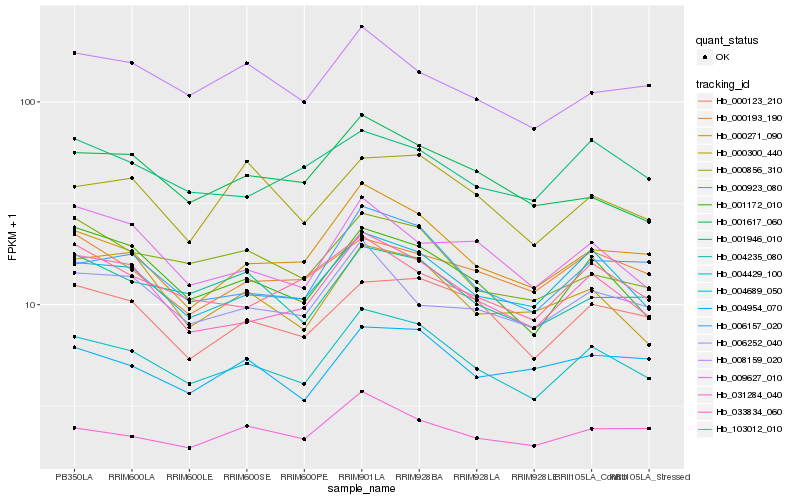
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004429_100 |
0.0 |
- |
- |
PREDICTED: asparagine synthetase domain-containing protein 1 [Jatropha curcas] |
| 2 |
Hb_004689_050 |
0.0539628473 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_033834_060 |
0.0591720971 |
- |
- |
DNA-binding bromodomain-containing family protein [Populus trichocarpa] |
| 4 |
Hb_000271_090 |
0.06433413 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 5 |
Hb_006157_020 |
0.065279561 |
- |
- |
PREDICTED: nucleolin 1-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_001172_010 |
0.0659151801 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 7 |
Hb_000123_210 |
0.0704371623 |
- |
- |
PREDICTED: uncharacterized protein LOC105641576 isoform X2 [Jatropha curcas] |
| 8 |
Hb_004235_080 |
0.0729860983 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 9 |
Hb_000923_080 |
0.0773035622 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2A [Jatropha curcas] |
| 10 |
Hb_001617_060 |
0.0789192434 |
- |
- |
PREDICTED: polyadenylate-binding protein RBP45B isoform X1 [Jatropha curcas] |
| 11 |
Hb_031284_040 |
0.0821882411 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g30700 [Jatropha curcas] |
| 12 |
Hb_004954_070 |
0.0821952496 |
- |
- |
PREDICTED: uncharacterized protein LOC105645187 [Jatropha curcas] |
| 13 |
Hb_008159_020 |
0.083519103 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 14 |
Hb_001946_010 |
0.0836341557 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 15 |
Hb_103012_010 |
0.0837851398 |
- |
- |
PREDICTED: uncharacterized protein LOC105638878 [Jatropha curcas] |
| 16 |
Hb_000300_440 |
0.0838065043 |
- |
- |
MIF4G domain and MA3 domain-containing protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_009627_010 |
0.0840752988 |
- |
- |
Protein dom-3, putative [Ricinus communis] |
| 18 |
Hb_000856_310 |
0.0857624224 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Jatropha curcas] |
| 19 |
Hb_000193_190 |
0.086265662 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 16 [Jatropha curcas] |
| 20 |
Hb_006252_040 |
0.0864397748 |
- |
- |
queuine tRNA-ribosyltransferase, putative [Ricinus communis] |