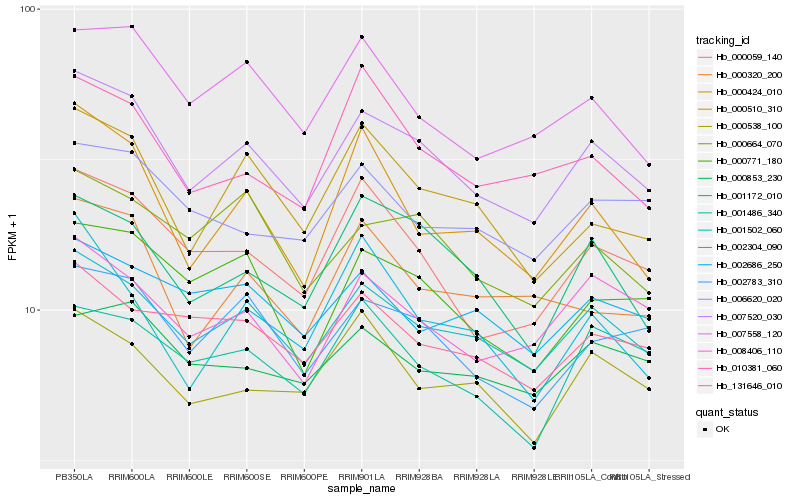
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007520_030 |
0.0 |
- |
- |
PREDICTED: peroxisomal membrane protein PEX14 [Jatropha curcas] |
| 2 |
Hb_002783_310 |
0.0612840967 |
- |
- |
PREDICTED: tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit trm6 [Jatropha curcas] |
| 3 |
Hb_000059_140 |
0.0683716844 |
- |
- |
rnase h, putative [Ricinus communis] |
| 4 |
Hb_008406_110 |
0.0697855825 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a isoform X1 [Jatropha curcas] |
| 5 |
Hb_001172_010 |
0.0709061407 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 6 |
Hb_000771_180 |
0.0713585684 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 7 |
Hb_001502_060 |
0.0713858933 |
- |
- |
DNA mismatch repair protein MSH2, putative [Ricinus communis] |
| 8 |
Hb_007558_120 |
0.0727150505 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 9 |
Hb_000664_070 |
0.0727376976 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 10 |
Hb_000510_310 |
0.073146107 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 11 |
Hb_131646_010 |
0.0752385169 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000538_100 |
0.0769203849 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002304_090 |
0.077927697 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX35 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000853_230 |
0.0785530838 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 15 |
Hb_006620_020 |
0.079008579 |
- |
- |
PREDICTED: uncharacterized protein LOC105638878 [Jatropha curcas] |
| 16 |
Hb_010381_060 |
0.0793938011 |
- |
- |
DEAD-box ATP-dependent RNA helicase 20 -like protein [Gossypium arboreum] |
| 17 |
Hb_002686_250 |
0.0796676537 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 65 kDa protein isoform X1 [Jatropha curcas] |
| 18 |
Hb_000320_200 |
0.0797225419 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 6 homolog [Jatropha curcas] |
| 19 |
Hb_001486_340 |
0.0799670643 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 1-like [Jatropha curcas] |
| 20 |
Hb_000424_010 |
0.0819748989 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |