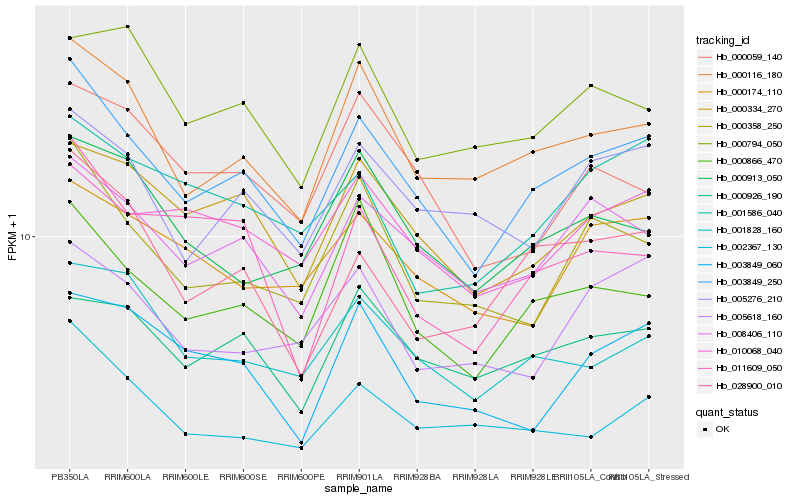
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003849_250 |
0.0 |
- |
- |
PREDICTED: protein MAK16 homolog [Jatropha curcas] |
| 2 |
Hb_008406_110 |
0.0680606832 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a isoform X1 [Jatropha curcas] |
| 3 |
Hb_000116_180 |
0.0702690499 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein H isoform X1 [Jatropha curcas] |
| 4 |
Hb_000334_270 |
0.0727536671 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000866_470 |
0.0749046055 |
- |
- |
PREDICTED: uncharacterized protein LOC105643015 [Jatropha curcas] |
| 6 |
Hb_000358_250 |
0.0828042504 |
- |
- |
PREDICTED: cell division cycle protein 27 homolog B [Jatropha curcas] |
| 7 |
Hb_001828_160 |
0.0848785883 |
- |
- |
hypothetical protein F775_31105 [Aegilops tauschii] |
| 8 |
Hb_005276_210 |
0.0876355733 |
- |
- |
PREDICTED: DNA repair helicase UVH6 isoform X2 [Jatropha curcas] |
| 9 |
Hb_010068_040 |
0.0888485978 |
- |
- |
PREDICTED: uncharacterized protein LOC105632964 [Jatropha curcas] |
| 10 |
Hb_000926_190 |
0.0888614613 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 3, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_028900_010 |
0.089295252 |
- |
- |
PREDICTED: uncharacterized protein LOC105631404 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002367_130 |
0.0911814808 |
- |
- |
UNC-50 family protein isoform 1 [Theobroma cacao] |
| 13 |
Hb_000913_050 |
0.0913217848 |
- |
- |
hypothetical protein JCGZ_17329 [Jatropha curcas] |
| 14 |
Hb_000174_110 |
0.0922876237 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 15 |
Hb_001586_040 |
0.0957482229 |
- |
- |
hypothetical protein JCGZ_17187 [Jatropha curcas] |
| 16 |
Hb_000059_140 |
0.0969717509 |
- |
- |
rnase h, putative [Ricinus communis] |
| 17 |
Hb_005618_160 |
0.0972974846 |
- |
- |
PREDICTED: putative proline--tRNA ligase C19C7.06 [Jatropha curcas] |
| 18 |
Hb_003849_060 |
0.0981951581 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 19 |
Hb_011609_050 |
0.0983306741 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000794_050 |
0.0989660124 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 12 [Jatropha curcas] |