| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
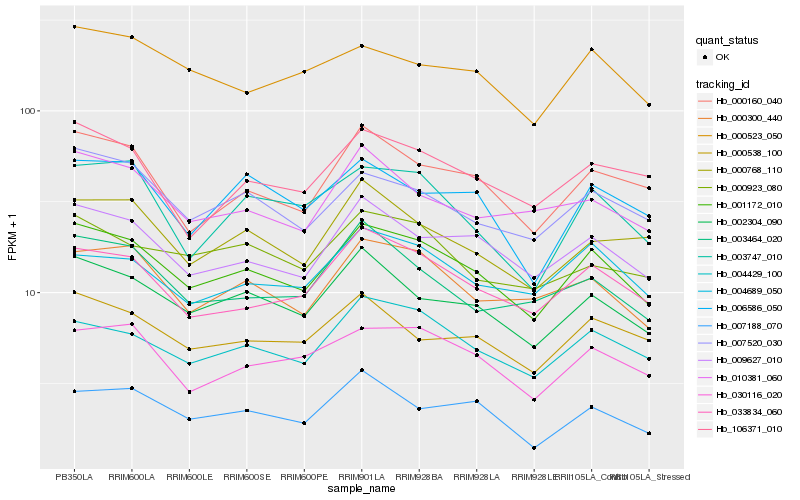
Hb_001172_010 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 2 |
Hb_009627_010 |
0.0607542445 |
- |
- |
Protein dom-3, putative [Ricinus communis] |
| 3 |
Hb_002304_090 |
0.0612167015 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX35 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004429_100 |
0.0659151801 |
- |
- |
PREDICTED: asparagine synthetase domain-containing protein 1 [Jatropha curcas] |
| 5 |
Hb_033834_060 |
0.0666679936 |
- |
- |
DNA-binding bromodomain-containing family protein [Populus trichocarpa] |
| 6 |
Hb_007520_030 |
0.0709061407 |
- |
- |
PREDICTED: peroxisomal membrane protein PEX14 [Jatropha curcas] |
| 7 |
Hb_000160_040 |
0.0713814021 |
- |
- |
PREDICTED: methyltransferase-like protein 10 [Jatropha curcas] |
| 8 |
Hb_000300_440 |
0.0723588583 |
- |
- |
MIF4G domain and MA3 domain-containing protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_030116_020 |
0.0725181938 |
- |
- |
hypothetical protein POPTR_0016s13630g [Populus trichocarpa] |
| 10 |
Hb_000538_100 |
0.0793131636 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000523_050 |
0.0814672952 |
- |
- |
microsomal glutathione s-transferase, putative [Ricinus communis] |
| 12 |
Hb_003747_010 |
0.0814787627 |
- |
- |
hypothetical protein JCGZ_24080 [Jatropha curcas] |
| 13 |
Hb_004689_050 |
0.0818682087 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_106371_010 |
0.0819088771 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 15 |
Hb_007188_070 |
0.083097729 |
- |
- |
prokaryotic DNA topoisomerase, putative [Ricinus communis] |
| 16 |
Hb_006586_050 |
0.0847521374 |
- |
- |
PREDICTED: probable protein phosphatase 2C 39 [Jatropha curcas] |
| 17 |
Hb_000923_080 |
0.0851369014 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2A [Jatropha curcas] |
| 18 |
Hb_010381_060 |
0.0854311838 |
- |
- |
DEAD-box ATP-dependent RNA helicase 20 -like protein [Gossypium arboreum] |
| 19 |
Hb_000768_110 |
0.085972947 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003464_020 |
0.0860583425 |
- |
- |
srpk, putative [Ricinus communis] |