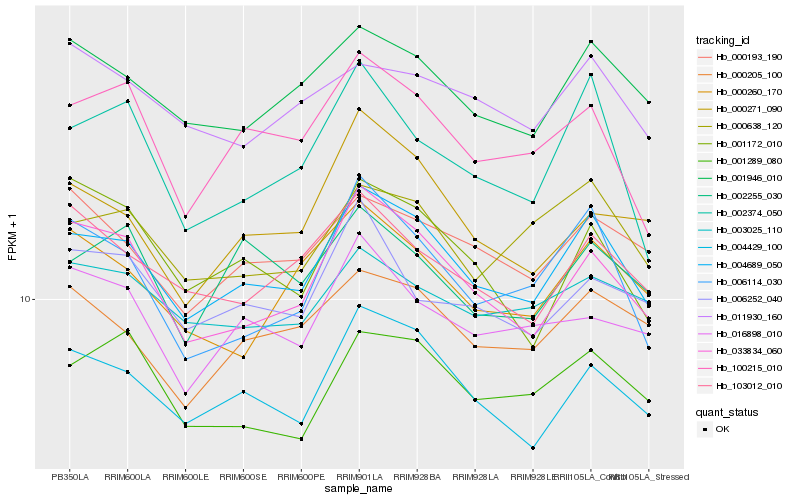
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004689_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004429_100 |
0.0539628473 |
- |
- |
PREDICTED: asparagine synthetase domain-containing protein 1 [Jatropha curcas] |
| 3 |
Hb_033834_060 |
0.0580862539 |
- |
- |
DNA-binding bromodomain-containing family protein [Populus trichocarpa] |
| 4 |
Hb_001946_010 |
0.0604625644 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 5 |
Hb_000638_120 |
0.0694986714 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit alpha 2, mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_000205_100 |
0.0696980719 |
- |
- |
PREDICTED: exportin-4 [Jatropha curcas] |
| 7 |
Hb_100215_010 |
0.0699681988 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein R isoform X1 [Jatropha curcas] |
| 8 |
Hb_000193_190 |
0.0726646029 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 16 [Jatropha curcas] |
| 9 |
Hb_001289_080 |
0.0726819885 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 10 |
Hb_006114_030 |
0.0727044528 |
transcription factor |
TF Family: SET |
hypothetical protein CISIN_1g011639mg [Citrus sinensis] |
| 11 |
Hb_103012_010 |
0.0736120233 |
- |
- |
PREDICTED: uncharacterized protein LOC105638878 [Jatropha curcas] |
| 12 |
Hb_000260_170 |
0.076852467 |
- |
- |
PREDICTED: putative zinc transporter At3g08650 [Cucumis sativus] |
| 13 |
Hb_003025_110 |
0.0781848929 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002374_050 |
0.0788792123 |
- |
- |
PREDICTED: auxilin-related protein 1 [Jatropha curcas] |
| 15 |
Hb_011930_160 |
0.0802313636 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 38-like [Jatropha curcas] |
| 16 |
Hb_016898_010 |
0.0816334288 |
- |
- |
PREDICTED: uncharacterized protein PFB0145c [Jatropha curcas] |
| 17 |
Hb_000271_090 |
0.0816967281 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 18 |
Hb_001172_010 |
0.0818682087 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 19 |
Hb_002255_030 |
0.0826536989 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 20 |
Hb_006252_040 |
0.0827924807 |
- |
- |
queuine tRNA-ribosyltransferase, putative [Ricinus communis] |