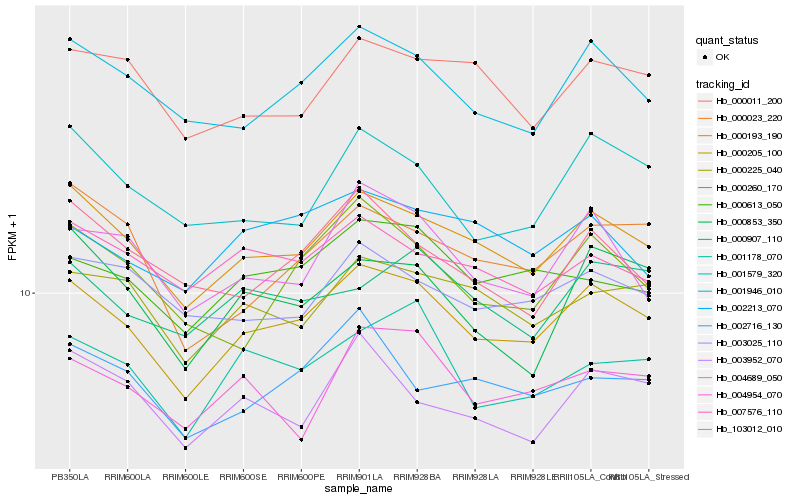
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000205_100 |
0.0 |
- |
- |
PREDICTED: exportin-4 [Jatropha curcas] |
| 2 |
Hb_000193_190 |
0.0409710556 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 16 [Jatropha curcas] |
| 3 |
Hb_001946_010 |
0.0586577574 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 4 |
Hb_000225_040 |
0.0690222435 |
- |
- |
PREDICTED: translation factor GUF1 homolog, mitochondrial [Nelumbo nucifera] |
| 5 |
Hb_000613_050 |
0.0690970715 |
- |
- |
PREDICTED: uncharacterized protein LOC105641544 isoform X2 [Jatropha curcas] |
| 6 |
Hb_004689_050 |
0.0696980719 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001178_070 |
0.0734937658 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein FBL11 [Jatropha curcas] |
| 8 |
Hb_001579_320 |
0.0739525504 |
- |
- |
pyroglutamyl-peptidase I, putative [Ricinus communis] |
| 9 |
Hb_000260_170 |
0.0743199344 |
- |
- |
PREDICTED: putative zinc transporter At3g08650 [Cucumis sativus] |
| 10 |
Hb_000011_200 |
0.0749054781 |
- |
- |
PREDICTED: 26S protease regulatory subunit 7 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000023_220 |
0.0754690185 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 18 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003025_110 |
0.0772522995 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007576_110 |
0.0784538267 |
- |
- |
Endoplasmic reticulum vesicle transporter protein [Theobroma cacao] |
| 14 |
Hb_002213_070 |
0.0784816604 |
- |
- |
epsilon-adaptin family protein [Populus trichocarpa] |
| 15 |
Hb_000853_350 |
0.0784826591 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like [Jatropha curcas] |
| 16 |
Hb_003952_070 |
0.0792225581 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 17 |
Hb_004954_070 |
0.0795973128 |
- |
- |
PREDICTED: uncharacterized protein LOC105645187 [Jatropha curcas] |
| 18 |
Hb_103012_010 |
0.0799892744 |
- |
- |
PREDICTED: uncharacterized protein LOC105638878 [Jatropha curcas] |
| 19 |
Hb_000907_110 |
0.0800998391 |
- |
- |
dihydroxyacetone kinase family protein [Populus trichocarpa] |
| 20 |
Hb_002716_130 |
0.0804818379 |
- |
- |
PREDICTED: uncharacterized protein LOC105645470 isoform X2 [Jatropha curcas] |