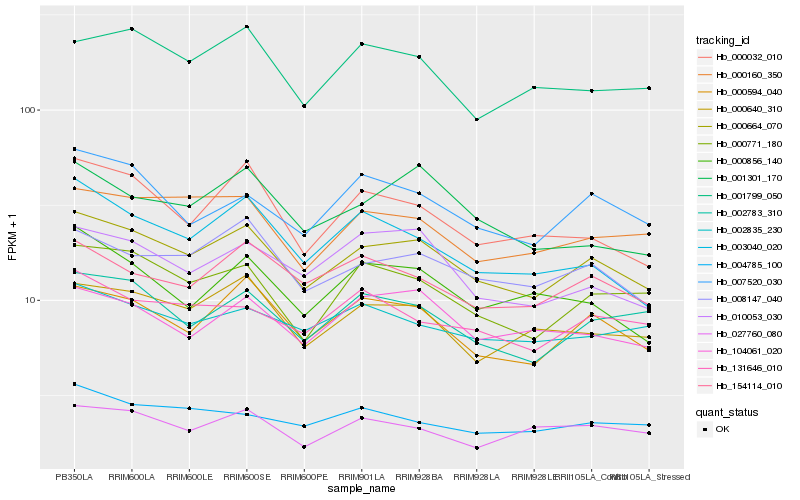
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000664_070 |
0.0 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 2 |
Hb_000594_040 |
0.0572836894 |
- |
- |
hypothetical protein L484_013922 [Morus notabilis] |
| 3 |
Hb_008147_040 |
0.0619096852 |
- |
- |
PREDICTED: protein FLX-like 2 [Jatropha curcas] |
| 4 |
Hb_000771_180 |
0.0668451073 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 5 |
Hb_000640_310 |
0.0689667017 |
- |
- |
hypothetical protein JCGZ_14055 [Jatropha curcas] |
| 6 |
Hb_002783_310 |
0.0692606548 |
- |
- |
PREDICTED: tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit trm6 [Jatropha curcas] |
| 7 |
Hb_000160_350 |
0.0715672783 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 4 isoform X3 [Vitis vinifera] |
| 8 |
Hb_154114_010 |
0.0720791283 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3A isoform X2 [Jatropha curcas] |
| 9 |
Hb_007520_030 |
0.0727376976 |
- |
- |
PREDICTED: peroxisomal membrane protein PEX14 [Jatropha curcas] |
| 10 |
Hb_104061_020 |
0.0751469342 |
- |
- |
PREDICTED: UV-stimulated scaffold protein A homolog [Jatropha curcas] |
| 11 |
Hb_010053_030 |
0.0759831781 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 12 |
Hb_000032_010 |
0.076157735 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 13 |
Hb_001799_050 |
0.0788182554 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 14 |
Hb_131646_010 |
0.079956544 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_002835_230 |
0.0812667299 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_003040_020 |
0.0818052418 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 17 |
Hb_001301_170 |
0.081950324 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 18 |
Hb_027760_080 |
0.082397137 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_004785_100 |
0.0832488032 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000856_140 |
0.0834717167 |
- |
- |
PREDICTED: uncharacterized protein LOC105640478 isoform X1 [Jatropha curcas] |