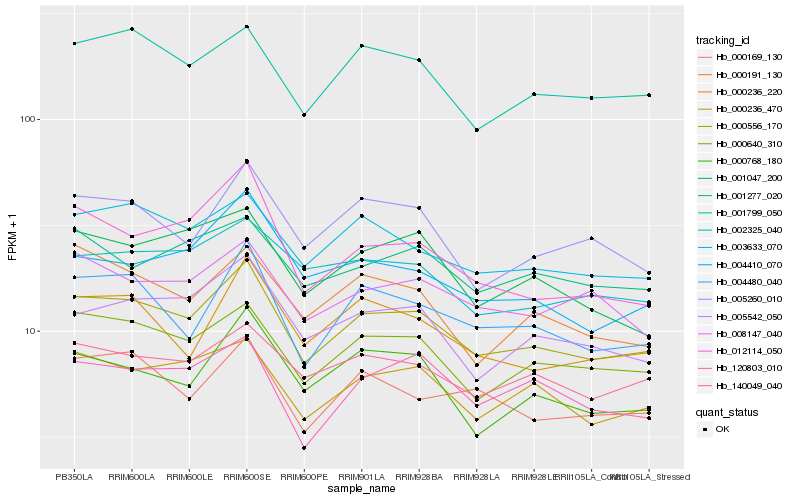
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000556_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000640_310 |
0.0525301074 |
- |
- |
hypothetical protein JCGZ_14055 [Jatropha curcas] |
| 3 |
Hb_000236_470 |
0.0622554948 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55840 [Jatropha curcas] |
| 4 |
Hb_003633_070 |
0.0625572807 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 5 |
Hb_001799_050 |
0.0631977002 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 6 |
Hb_140049_040 |
0.0646379193 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 7 |
Hb_004480_040 |
0.0663427419 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_005542_050 |
0.0670626144 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable inactive serine/threonine-protein kinase scy1 [Prunus mume] |
| 9 |
Hb_005260_010 |
0.0710959465 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 10 |
Hb_008147_040 |
0.0738275982 |
- |
- |
PREDICTED: protein FLX-like 2 [Jatropha curcas] |
| 11 |
Hb_000236_220 |
0.0738920332 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 12 |
Hb_002325_040 |
0.0747711522 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000768_180 |
0.075216287 |
- |
- |
PREDICTED: uncharacterized protein LOC105632528 [Jatropha curcas] |
| 14 |
Hb_000169_130 |
0.0760974487 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 15 |
Hb_120803_010 |
0.0766464257 |
- |
- |
PREDICTED: protein kinase and PP2C-like domain-containing protein [Malus domestica] |
| 16 |
Hb_001277_020 |
0.0770411448 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 17 |
Hb_004410_070 |
0.0779980352 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 18 |
Hb_000191_130 |
0.0795386017 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 19 |
Hb_012114_050 |
0.0795945034 |
- |
- |
PREDICTED: uncharacterized protein LOC105645815 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001047_200 |
0.0795960019 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |