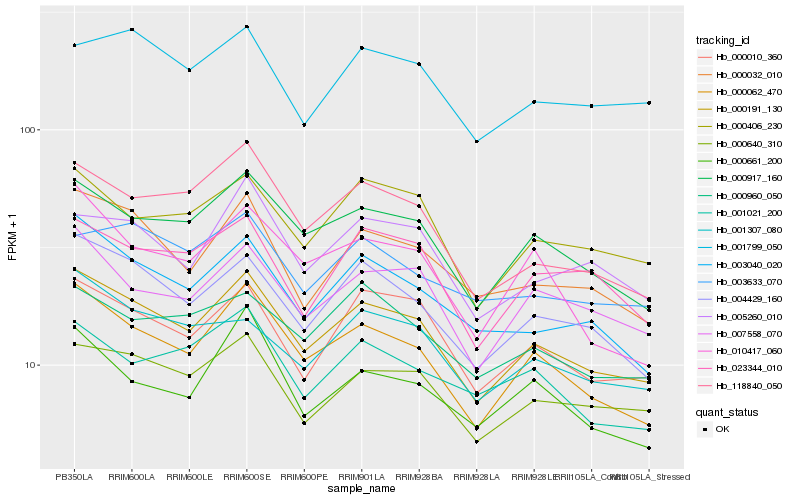
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000191_130 |
0.0 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 2 |
Hb_000062_470 |
0.0421529316 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 3 |
Hb_000917_160 |
0.0476507075 |
- |
- |
PREDICTED: protein decapping 5 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004429_160 |
0.0492670812 |
- |
- |
PREDICTED: serine/threonine-protein kinase TAO3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000010_360 |
0.0560400341 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 6 |
Hb_000640_310 |
0.0576859713 |
- |
- |
hypothetical protein JCGZ_14055 [Jatropha curcas] |
| 7 |
Hb_000032_010 |
0.0604170895 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 8 |
Hb_000406_230 |
0.0607478067 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 9 |
Hb_007558_070 |
0.0609480808 |
- |
- |
PREDICTED: pre-mRNA-processing factor 39 isoform X2 [Jatropha curcas] |
| 10 |
Hb_003040_020 |
0.0638696995 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 11 |
Hb_118840_050 |
0.0656547223 |
- |
- |
hypothetical protein CISIN_1g0019781mg, partial [Citrus sinensis] |
| 12 |
Hb_001307_080 |
0.0674682438 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 13 |
Hb_005260_010 |
0.06973725 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 14 |
Hb_023344_010 |
0.0702096933 |
- |
- |
PREDICTED: pre-mRNA-processing factor 39 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000960_050 |
0.0719949752 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 16 |
Hb_001799_050 |
0.0752489793 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 17 |
Hb_000661_200 |
0.0756405141 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001021_200 |
0.0779638503 |
desease resistance |
Gene Name: AAA |
PREDICTED: peroxisome biogenesis protein 6 [Jatropha curcas] |
| 19 |
Hb_010417_060 |
0.0780427205 |
- |
- |
PREDICTED: protein SGT1 homolog A-like [Jatropha curcas] |
| 20 |
Hb_003633_070 |
0.0793991944 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |