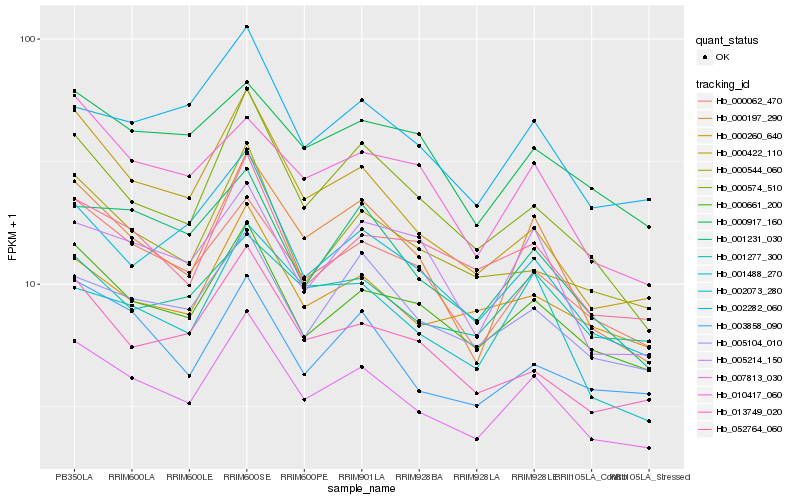
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007813_030 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At4g19050 [Jatropha curcas] |
| 2 |
Hb_000661_200 |
0.061254195 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000062_470 |
0.0680492116 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 4 |
Hb_000197_290 |
0.070361864 |
- |
- |
PREDICTED: A-agglutinin anchorage subunit [Jatropha curcas] |
| 5 |
Hb_000574_510 |
0.0797653505 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 6 |
Hb_052764_060 |
0.080600467 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 7 |
Hb_005104_010 |
0.0819639133 |
- |
- |
PREDICTED: uncharacterized protein LOC105140796 isoform X1 [Populus euphratica] |
| 8 |
Hb_000544_060 |
0.0823663474 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 9 |
Hb_001231_030 |
0.0825280503 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 10 |
Hb_002282_060 |
0.0826151868 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_000260_640 |
0.0846056396 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 12 |
Hb_003858_090 |
0.0849833933 |
- |
- |
Filamin-A-interacting protein, putative [Ricinus communis] |
| 13 |
Hb_002073_280 |
0.0864488314 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000422_110 |
0.0875988904 |
- |
- |
PREDICTED: uncharacterized protein At1g08160-like [Jatropha curcas] |
| 15 |
Hb_013749_020 |
0.0899165713 |
- |
- |
PREDICTED: inositol phosphorylceramide glucuronosyltransferase 1 [Jatropha curcas] |
| 16 |
Hb_005214_150 |
0.0906801258 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 17 |
Hb_001277_300 |
0.0906803638 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 18 |
Hb_000917_160 |
0.0912648934 |
- |
- |
PREDICTED: protein decapping 5 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001488_270 |
0.0913963197 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 20 |
Hb_010417_060 |
0.0918647741 |
- |
- |
PREDICTED: protein SGT1 homolog A-like [Jatropha curcas] |